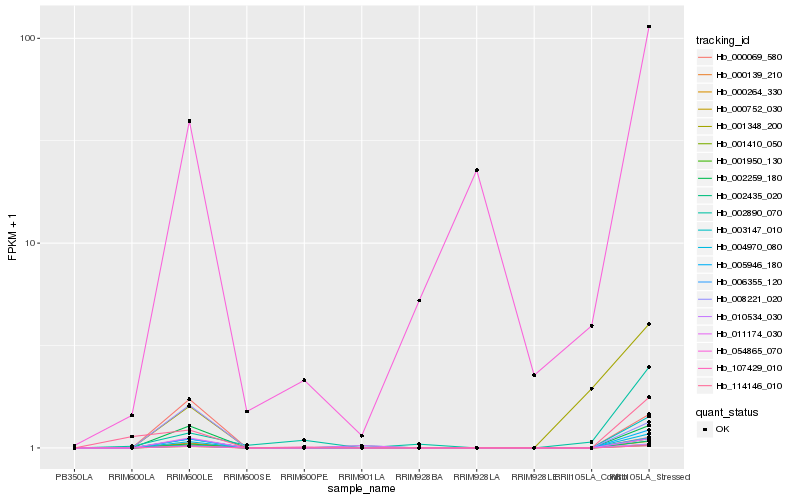
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001950_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_010534_030 |
0.0012547298 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_000752_030 |
0.0016707874 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0978110 [Ricinus communis] |
| 4 |
Hb_004970_080 |
0.0023721213 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 5 |
Hb_000264_330 |
0.0112804738 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 6 |
Hb_000139_210 |
0.0139158187 |
- |
- |
- |
| 7 |
Hb_005946_180 |
0.0225240206 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |
| 8 |
Hb_001410_050 |
0.1509741225 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 9 |
Hb_006355_120 |
0.1559755185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002259_180 |
0.1740571598 |
- |
- |
- |
| 11 |
Hb_002435_020 |
0.2363136157 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 12 |
Hb_114146_010 |
0.259825673 |
- |
- |
hypothetical protein JCGZ_15608 [Jatropha curcas] |
| 13 |
Hb_000069_580 |
0.2743900468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003147_010 |
0.2927976336 |
- |
- |
PREDICTED: bark storage protein A-like [Jatropha curcas] |
| 15 |
Hb_011174_030 |
0.3017287991 |
- |
- |
- |
| 16 |
Hb_008221_020 |
0.3025433807 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 17 |
Hb_054865_070 |
0.3093646933 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 18 |
Hb_002890_070 |
0.3237285825 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 19 |
Hb_107429_010 |
0.3427866025 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 20 |
Hb_001348_200 |
0.3568898843 |
- |
- |
- |