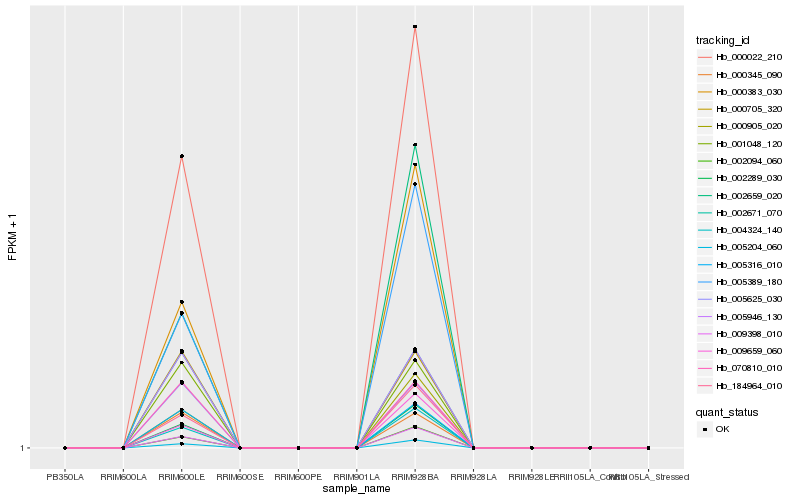
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638395 [Jatropha curcas] |
| 2 |
Hb_009398_010 |
0.035228918 |
- |
- |
Putative pectate lyase 2 [Glycine soja] |
| 3 |
Hb_002289_030 |
0.0365333439 |
- |
- |
PREDICTED: uncharacterized protein LOC103957017 [Pyrus x bretschneideri] |
| 4 |
Hb_005204_060 |
0.0448639817 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_002094_060 |
0.0460789451 |
- |
- |
- |
| 6 |
Hb_184964_010 |
0.0470444653 |
- |
- |
- |
| 7 |
Hb_004324_140 |
0.0477665494 |
- |
- |
PREDICTED: uncharacterized protein LOC105648358 [Jatropha curcas] |
| 8 |
Hb_000905_020 |
0.0509326602 |
- |
- |
PREDICTED: cytochrome P450 93A2-like [Jatropha curcas] |
| 9 |
Hb_005316_010 |
0.0573255749 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000383_030 |
0.0604790546 |
- |
- |
- |
| 11 |
Hb_005389_180 |
0.0624176083 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 12 |
Hb_002671_070 |
0.0825554933 |
- |
- |
hypothetical protein POPTR_0006s05220g, partial [Populus trichocarpa] |
| 13 |
Hb_005946_130 |
0.0825554933 |
- |
- |
- |
| 14 |
Hb_001048_120 |
0.0843881621 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_070810_010 |
0.0880848369 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 16 |
Hb_005625_030 |
0.088253477 |
- |
- |
ribosomal protein S12 (mitochondrion) [Chrysobalanus icaco] |
| 17 |
Hb_009659_060 |
0.0901420647 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 18 |
Hb_000705_320 |
0.0912998087 |
- |
- |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 19 |
Hb_002659_020 |
0.0921668757 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 20 |
Hb_000345_090 |
0.0939812617 |
- |
- |
Cell cycle checkpoint control protein family isoform 5 [Theobroma cacao] |