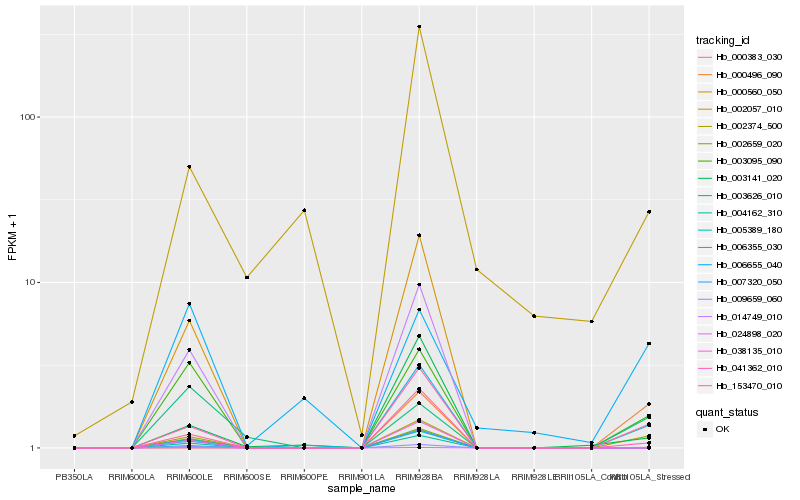
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_500 |
0.0 |
- |
- |
PREDICTED: putative F-box protein PP2-B12 [Gossypium raimondii] |
| 2 |
Hb_153470_010 |
0.1133512335 |
- |
- |
PREDICTED: uncharacterized protein LOC104804963 [Tarenaya hassleriana] |
| 3 |
Hb_003095_090 |
0.2032721815 |
- |
- |
hypothetical protein JCGZ_10029 [Jatropha curcas] |
| 4 |
Hb_024898_020 |
0.2188596354 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_041362_010 |
0.2819183838 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 6 |
Hb_006655_040 |
0.2997276727 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 7 |
Hb_003626_010 |
0.3161647912 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 8 |
Hb_006355_030 |
0.3216346768 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 9 |
Hb_003141_020 |
0.3220809377 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 10 |
Hb_002057_010 |
0.3438369328 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_007320_050 |
0.3464635702 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 12 |
Hb_004162_310 |
0.346494131 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 13 |
Hb_014749_010 |
0.3465606422 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 14 |
Hb_002659_020 |
0.3476067884 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 15 |
Hb_009659_060 |
0.3477659321 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 16 |
Hb_000560_050 |
0.3494673276 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 17 |
Hb_005389_180 |
0.3509630647 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 18 |
Hb_000496_090 |
0.3511190994 |
- |
- |
PREDICTED: inhibitor of trypsin and hageman factor-like [Gossypium raimondii] |
| 19 |
Hb_038135_010 |
0.3512032403 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 20 |
Hb_000383_030 |
0.3512565531 |
- |
- |
- |