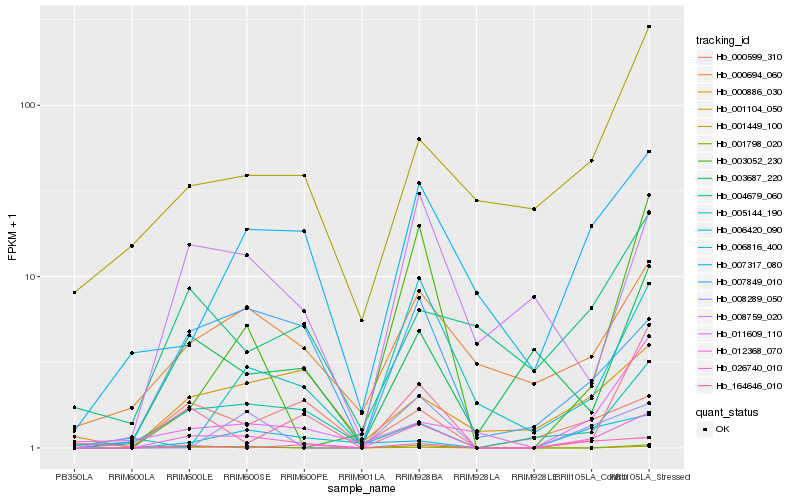
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012368_070 |
0.0 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001798_020 |
0.2670345077 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 3 |
Hb_007849_010 |
0.2873165604 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_001104_050 |
0.2894822242 |
- |
- |
- |
| 5 |
Hb_005144_190 |
0.2895526226 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 6 |
Hb_007317_080 |
0.2902098726 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 7 |
Hb_000694_060 |
0.2905325554 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 8 |
Hb_003052_230 |
0.2908541955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_164646_010 |
0.292107787 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 10 |
Hb_008759_020 |
0.3030808517 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 11 |
Hb_003687_220 |
0.303255418 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 12 |
Hb_000599_310 |
0.3113386495 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 13 |
Hb_001449_100 |
0.3227822568 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 14 |
Hb_006420_090 |
0.3247250394 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 15 |
Hb_006816_400 |
0.3250623614 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 16 |
Hb_000886_030 |
0.3252240652 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 17 |
Hb_004679_060 |
0.325978992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008289_050 |
0.3277620618 |
- |
- |
hypothetical protein B456_001G045900 [Gossypium raimondii] |
| 19 |
Hb_026740_010 |
0.3291702858 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 20 |
Hb_011609_110 |
0.3294248703 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |