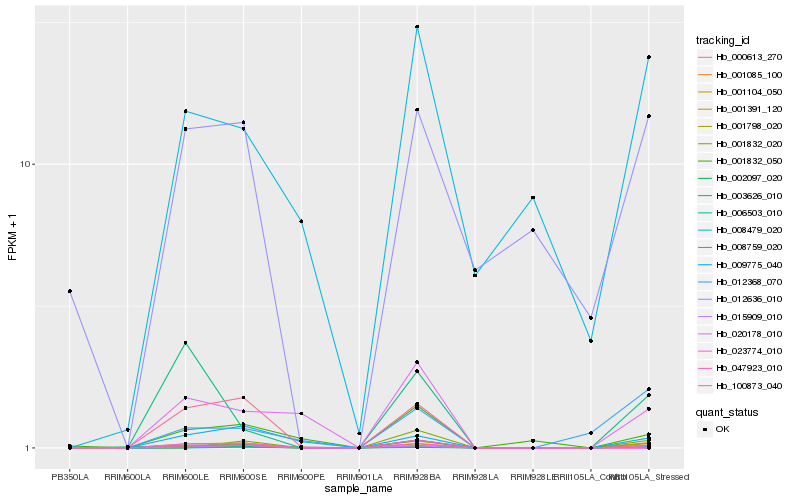
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001391_120 |
0.0 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 2 |
Hb_001798_020 |
0.1391207134 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 3 |
Hb_001104_050 |
0.2226569885 |
- |
- |
- |
| 4 |
Hb_002097_020 |
0.2320335999 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 5 |
Hb_020178_010 |
0.2801984964 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 6 |
Hb_009775_040 |
0.286897054 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 7 |
Hb_015909_010 |
0.2896053607 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_003626_010 |
0.298092773 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 9 |
Hb_001832_050 |
0.3003225354 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 10 |
Hb_008759_020 |
0.3163945911 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 11 |
Hb_001085_100 |
0.3168658089 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_100873_040 |
0.3209592935 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012636_010 |
0.3287919646 |
- |
- |
- |
| 14 |
Hb_012368_070 |
0.3296635221 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001832_020 |
0.3300702918 |
- |
- |
PREDICTED: uncharacterized protein LOC104584029 [Brachypodium distachyon] |
| 16 |
Hb_008479_020 |
0.3369631949 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 17 |
Hb_006503_010 |
0.3493995939 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_047923_010 |
0.3500803609 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 19 |
Hb_000613_270 |
0.3501576225 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 20 |
Hb_023774_010 |
0.3502916124 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |