| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
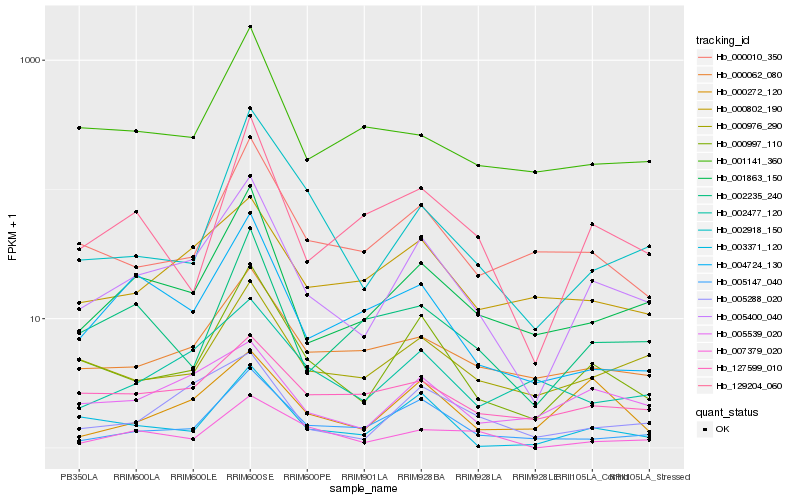
Hb_005400_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 2 |
Hb_000997_110 |
0.1278136139 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 3 |
Hb_001863_150 |
0.1439963844 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 4 |
Hb_005288_020 |
0.1551255735 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_005539_020 |
0.1720226073 |
- |
- |
- |
| 6 |
Hb_005147_040 |
0.1756540405 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 7 |
Hb_000976_290 |
0.1793617693 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 8 |
Hb_000062_080 |
0.1803351178 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 9 |
Hb_003371_120 |
0.1851564229 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 10 |
Hb_004724_130 |
0.1902808987 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 11 |
Hb_129204_060 |
0.1904873011 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 12 |
Hb_000802_190 |
0.192040354 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 13 |
Hb_002477_120 |
0.1944315441 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_127599_010 |
0.1952623753 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 15 |
Hb_000010_350 |
0.1953343859 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 16 |
Hb_002235_240 |
0.1967216858 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 17 |
Hb_007379_020 |
0.2001558728 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 18 |
Hb_000272_120 |
0.2026522209 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002918_150 |
0.2027070041 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 20 |
Hb_001141_360 |
0.2050444504 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |