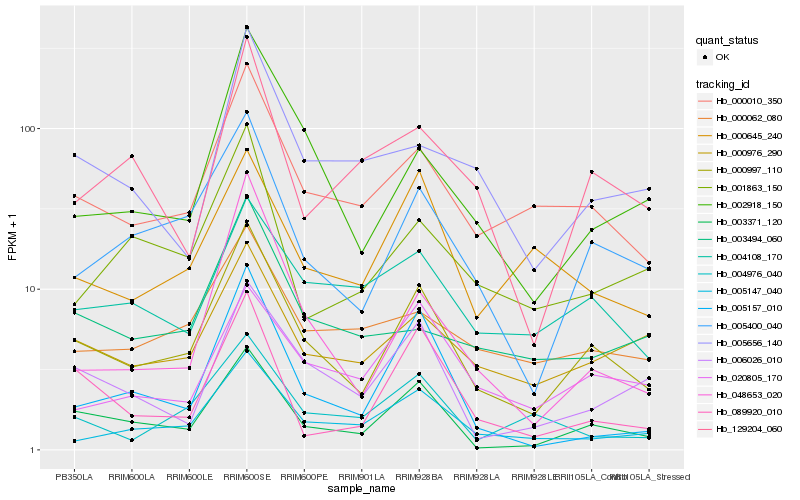
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 2 |
Hb_003371_120 |
0.1173751327 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 3 |
Hb_005400_040 |
0.1278136139 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 4 |
Hb_000010_350 |
0.1393340754 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 5 |
Hb_005147_040 |
0.1673073529 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 6 |
Hb_000976_290 |
0.170183654 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 7 |
Hb_002918_150 |
0.1719749322 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 8 |
Hb_006026_010 |
0.1770364954 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 3-like [Jatropha curcas] |
| 9 |
Hb_004108_170 |
0.1815705191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000062_080 |
0.1866698197 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 11 |
Hb_089920_010 |
0.1867648645 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_048653_020 |
0.1905114639 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_020805_170 |
0.1928431678 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 14 |
Hb_001863_150 |
0.1933632582 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 15 |
Hb_000645_240 |
0.194586997 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 16 |
Hb_005656_140 |
0.1947765797 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 17 |
Hb_003494_060 |
0.1952498592 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005157_010 |
0.1959591454 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 19 |
Hb_004976_040 |
0.1965363682 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 20 |
Hb_129204_060 |
0.1972853587 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |