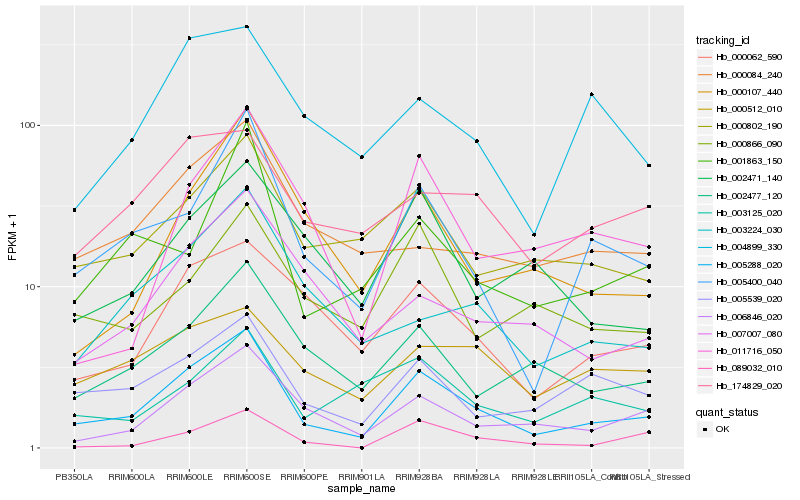
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_020 |
0.0 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_005400_040 |
0.1551255735 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 3 |
Hb_006846_020 |
0.1647443569 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 4 |
Hb_005539_020 |
0.1744720583 |
- |
- |
- |
| 5 |
Hb_000802_190 |
0.1799616363 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 6 |
Hb_000062_590 |
0.1812696365 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_002477_120 |
0.1831095087 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000107_440 |
0.1955379637 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 9 |
Hb_001863_150 |
0.1988069979 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 10 |
Hb_007007_080 |
0.1988367205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004899_330 |
0.2024366436 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 12 |
Hb_002471_140 |
0.2029103337 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 13 |
Hb_003224_030 |
0.2031865921 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 14 |
Hb_011716_050 |
0.2034183014 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 15 |
Hb_174829_020 |
0.2049168042 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 16 |
Hb_000866_090 |
0.2060330229 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_000084_240 |
0.2063856295 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 18 |
Hb_089032_010 |
0.2095517889 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 19 |
Hb_003125_020 |
0.2099464538 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_000512_010 |
0.2112528401 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |