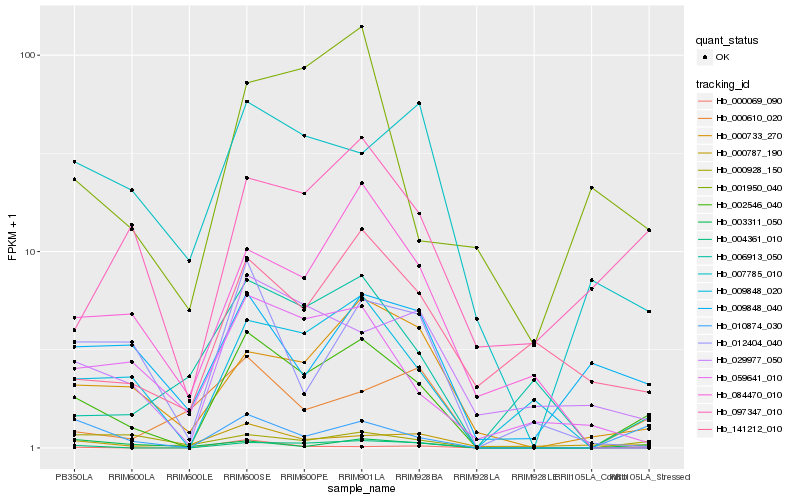
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002546_040 |
0.0 |
- |
- |
hypothetical protein CISIN_1g005599mg [Citrus sinensis] |
| 2 |
Hb_004361_010 |
0.2369467205 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 3 |
Hb_010874_030 |
0.2461972099 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 4 |
Hb_000733_270 |
0.2560858876 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 5 |
Hb_009848_020 |
0.2589452842 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 6 |
Hb_000787_190 |
0.2688280924 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 15-like [Jatropha curcas] |
| 7 |
Hb_029977_050 |
0.2725743224 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006913_050 |
0.2840051097 |
- |
- |
- |
| 9 |
Hb_059641_010 |
0.2843695075 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_084470_010 |
0.2879358618 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 11 |
Hb_012404_040 |
0.2896812604 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 12 |
Hb_097347_010 |
0.2947074555 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |
| 13 |
Hb_000069_090 |
0.2982392667 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 14 |
Hb_007785_010 |
0.2988202091 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 15 |
Hb_141212_010 |
0.2992405143 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_000610_020 |
0.299525808 |
- |
- |
unknown [Glycine max] |
| 17 |
Hb_000928_150 |
0.3008489849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003311_050 |
0.3023161528 |
- |
- |
PREDICTED: two-pore potassium channel 1-like [Jatropha curcas] |
| 19 |
Hb_009848_040 |
0.3046113343 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 20 |
Hb_001950_040 |
0.3051453653 |
- |
- |
- |