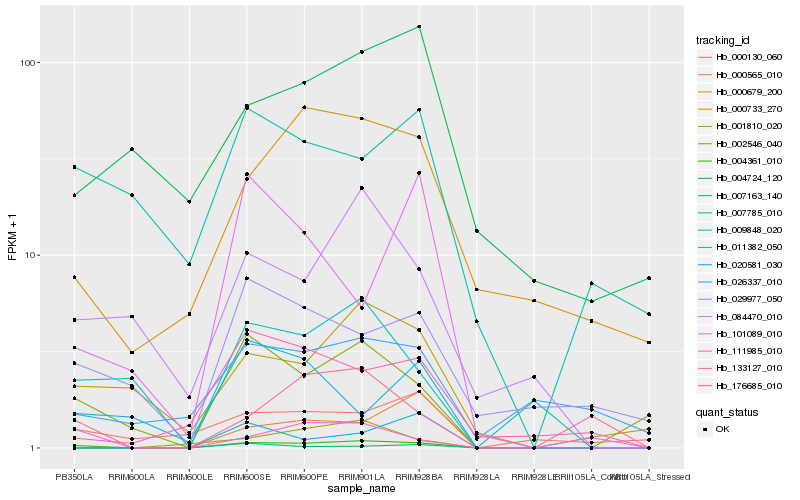
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004361_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 2 |
Hb_111985_010 |
0.2316951366 |
- |
- |
- |
| 3 |
Hb_002546_040 |
0.2369467205 |
- |
- |
hypothetical protein CISIN_1g005599mg [Citrus sinensis] |
| 4 |
Hb_000733_270 |
0.2747368899 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 5 |
Hb_009848_020 |
0.2851821764 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 6 |
Hb_084470_010 |
0.287418387 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_000679_200 |
0.2950377529 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 8 |
Hb_176685_010 |
0.2979791999 |
- |
- |
receptor-type protein kinase LRK1-like [Oryza sativa Japonica Group] |
| 9 |
Hb_133127_010 |
0.3073391519 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_029977_050 |
0.3189592024 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007163_140 |
0.3200228866 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_12606 [Jatropha curcas] |
| 12 |
Hb_000565_010 |
0.3248390537 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 13 |
Hb_011382_050 |
0.3293256408 |
- |
- |
hypothetical protein PRUPE_ppa025935mg [Prunus persica] |
| 14 |
Hb_000130_060 |
0.3303803125 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 15 |
Hb_001810_020 |
0.3318558319 |
- |
- |
- |
| 16 |
Hb_007785_010 |
0.3322045804 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 17 |
Hb_020581_030 |
0.3323731088 |
- |
- |
- |
| 18 |
Hb_026337_010 |
0.3333903444 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 19 |
Hb_004724_120 |
0.3362950234 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 20 |
Hb_101089_010 |
0.3363065484 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |