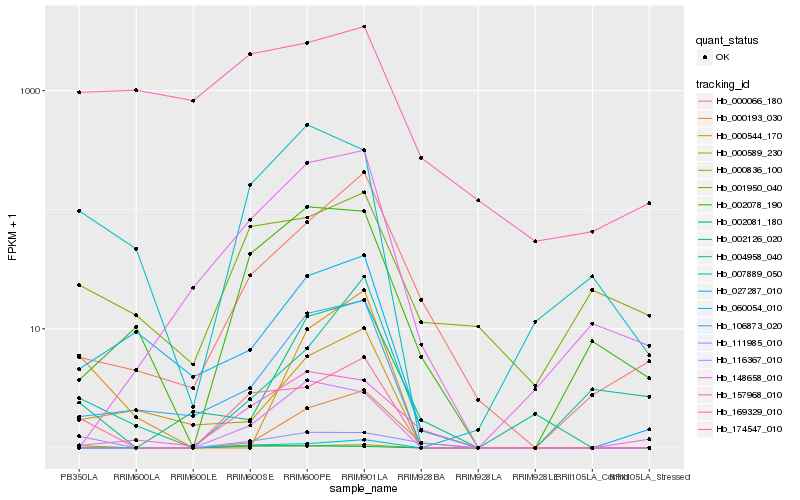
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174547_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000836_100 |
0.192727571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_027287_010 |
0.2072926148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_106873_020 |
0.2769770937 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 5 |
Hb_000066_180 |
0.2904392976 |
- |
- |
hypothetical protein POPTR_0008s02710g [Populus trichocarpa] |
| 6 |
Hb_007889_050 |
0.2999438457 |
- |
- |
Kynurenine formamidase [Gossypium arboreum] |
| 7 |
Hb_002078_190 |
0.3021505443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_148658_010 |
0.3054691569 |
- |
- |
- |
| 9 |
Hb_111985_010 |
0.3092352417 |
- |
- |
- |
| 10 |
Hb_000589_230 |
0.3105219283 |
- |
- |
unknown [Picea sitchensis] |
| 11 |
Hb_002081_180 |
0.3122422889 |
- |
- |
hypothetical protein PRUPE_ppa006101mg [Prunus persica] |
| 12 |
Hb_116367_010 |
0.3128688472 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Pyrus x bretschneideri] |
| 13 |
Hb_001950_040 |
0.316093841 |
- |
- |
- |
| 14 |
Hb_060054_010 |
0.3210249478 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 15 |
Hb_157968_010 |
0.33136324 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_002126_020 |
0.3342226068 |
- |
- |
ferrochelatase [Medicago truncatula] |
| 17 |
Hb_000193_030 |
0.3384894344 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 18 |
Hb_000544_170 |
0.3444962294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004958_040 |
0.3471650495 |
- |
- |
- |
| 20 |
Hb_169329_010 |
0.352713328 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |