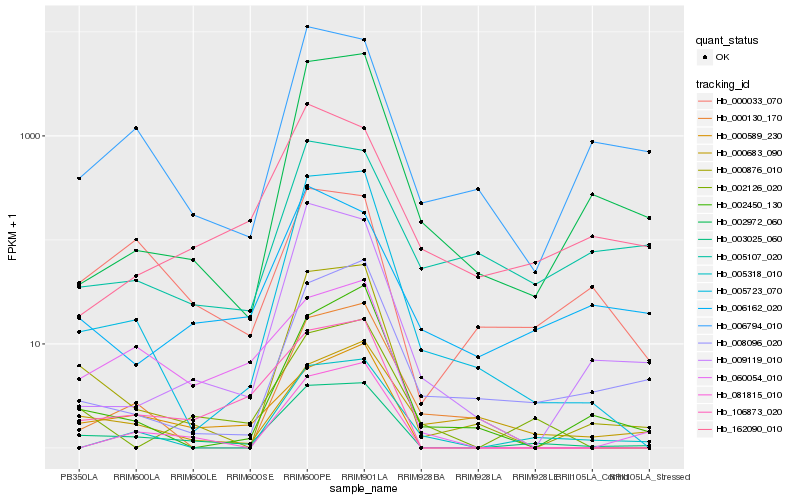
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003025_060 |
0.0 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 2 |
Hb_000876_010 |
0.1635100536 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 3 |
Hb_106873_020 |
0.1788992477 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_000589_230 |
0.1832827129 |
- |
- |
unknown [Picea sitchensis] |
| 5 |
Hb_005723_070 |
0.1884191724 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 6 |
Hb_002126_020 |
0.195433408 |
- |
- |
ferrochelatase [Medicago truncatula] |
| 7 |
Hb_060054_010 |
0.2013589961 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 8 |
Hb_006794_010 |
0.2086932106 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 9 |
Hb_000033_070 |
0.2132659967 |
- |
- |
unnamed protein product [Oryza sativa Japonica Group] |
| 10 |
Hb_081815_010 |
0.2165395109 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 11 |
Hb_002972_060 |
0.219783115 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_005318_010 |
0.2276585066 |
- |
- |
hypothetical protein POPTR_0012s15190g, partial [Populus trichocarpa] |
| 13 |
Hb_008096_020 |
0.2283533455 |
- |
- |
- |
| 14 |
Hb_009119_010 |
0.2299955867 |
- |
- |
- |
| 15 |
Hb_002450_130 |
0.230561781 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 16 |
Hb_006162_020 |
0.2360368507 |
- |
- |
- |
| 17 |
Hb_000683_090 |
0.2398099456 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 18 |
Hb_005107_020 |
0.2464937855 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 19 |
Hb_000130_170 |
0.2471151931 |
- |
- |
- |
| 20 |
Hb_162090_010 |
0.2471189897 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |