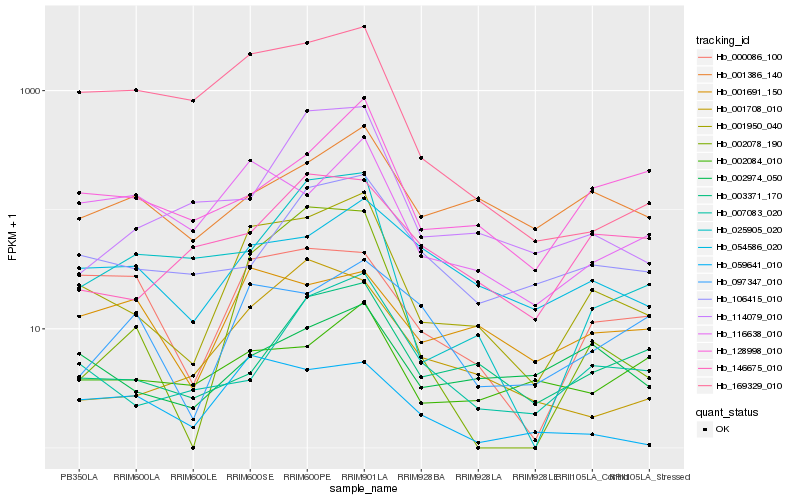
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001950_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_116638_010 |
0.2216627221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007083_020 |
0.2276804504 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 4 |
Hb_000086_100 |
0.2302484567 |
- |
- |
hypothetical protein POPTR_0005s24730g [Populus trichocarpa] |
| 5 |
Hb_002974_050 |
0.2321136244 |
- |
- |
- |
| 6 |
Hb_003371_170 |
0.2330572648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_169329_010 |
0.2333427567 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 8 |
Hb_054586_020 |
0.2351783502 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 9 |
Hb_128998_010 |
0.2364211733 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 10 |
Hb_106415_010 |
0.2376890589 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 11 |
Hb_025905_020 |
0.2419305565 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 12 |
Hb_001708_010 |
0.242423536 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 13 |
Hb_001691_150 |
0.2425858718 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Jatropha curcas] |
| 14 |
Hb_059641_010 |
0.2431706037 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_002078_190 |
0.2440422768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002084_010 |
0.2441610337 |
- |
- |
hypothetical protein VITISV_017990 [Vitis vinifera] |
| 17 |
Hb_001386_140 |
0.2484134761 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 18 |
Hb_114079_010 |
0.2499136611 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 19 |
Hb_146675_010 |
0.2499853237 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 20 |
Hb_097347_010 |
0.2500839068 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |