| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
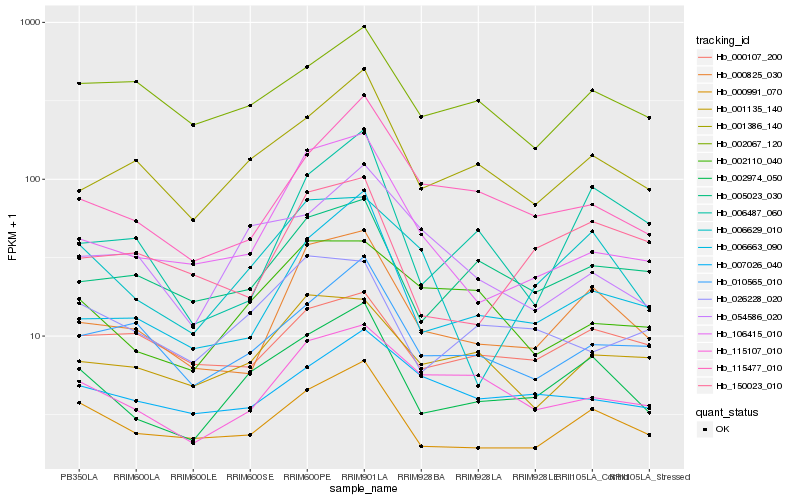
Hb_002974_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000991_070 |
0.1193305233 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 3 |
Hb_001386_140 |
0.1343203883 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_000825_030 |
0.1473007222 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_006663_090 |
0.1503975977 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 6 |
Hb_005023_030 |
0.1580667022 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 7 |
Hb_006629_010 |
0.1595440923 |
- |
- |
Glutathione S-transferase 2 isoform 3 [Theobroma cacao] |
| 8 |
Hb_010565_010 |
0.1601626594 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 9 |
Hb_115477_010 |
0.1641001636 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 10 |
Hb_115107_010 |
0.16757808 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 11 |
Hb_026228_020 |
0.1690920129 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 12 |
Hb_002067_120 |
0.1706508352 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 13 |
Hb_001135_140 |
0.174827702 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 14 |
Hb_002110_040 |
0.1767727489 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 15 |
Hb_054586_020 |
0.1770265968 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 16 |
Hb_106415_010 |
0.1790853938 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 17 |
Hb_007026_040 |
0.1818068786 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 18 |
Hb_006487_060 |
0.1824563446 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 19 |
Hb_000107_200 |
0.1843381456 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |
| 20 |
Hb_150023_010 |
0.1845604202 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |