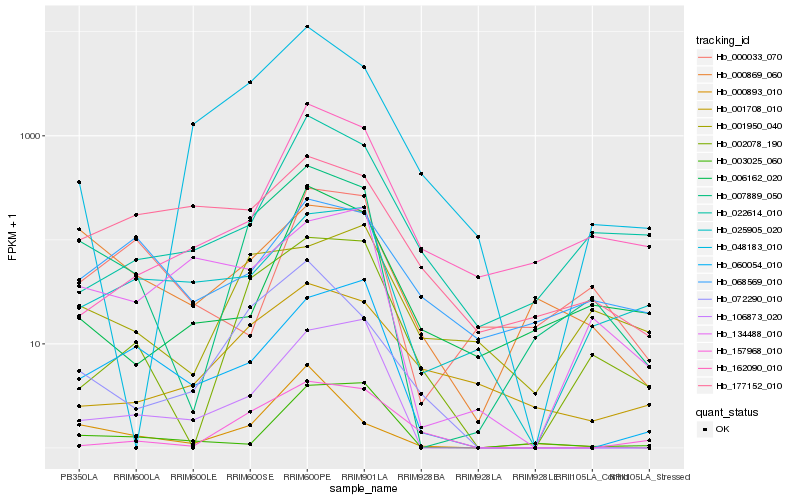
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007889_050 |
0.0 |
- |
- |
Kynurenine formamidase [Gossypium arboreum] |
| 2 |
Hb_002078_190 |
0.2110407806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000869_060 |
0.2349310325 |
- |
- |
hypothetical protein L484_003245 [Morus notabilis] |
| 4 |
Hb_000893_010 |
0.2621997352 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 5 |
Hb_072290_010 |
0.2625228799 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 6 |
Hb_106873_020 |
0.2733027328 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_001950_040 |
0.2758824836 |
- |
- |
- |
| 8 |
Hb_003025_060 |
0.2835295587 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 9 |
Hb_157968_010 |
0.2836421579 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_022614_010 |
0.2847504783 |
- |
- |
- |
| 11 |
Hb_001708_010 |
0.2848578177 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 12 |
Hb_060054_010 |
0.2878555318 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 13 |
Hb_006162_020 |
0.2886636181 |
- |
- |
- |
| 14 |
Hb_025905_020 |
0.2908907016 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 15 |
Hb_068569_010 |
0.2934816267 |
- |
- |
- |
| 16 |
Hb_134488_010 |
0.2958569748 |
- |
- |
- |
| 17 |
Hb_048183_010 |
0.2970808165 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_177152_010 |
0.2978467998 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 19 |
Hb_000033_070 |
0.2978727342 |
- |
- |
unnamed protein product [Oryza sativa Japonica Group] |
| 20 |
Hb_162090_010 |
0.2984241776 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |