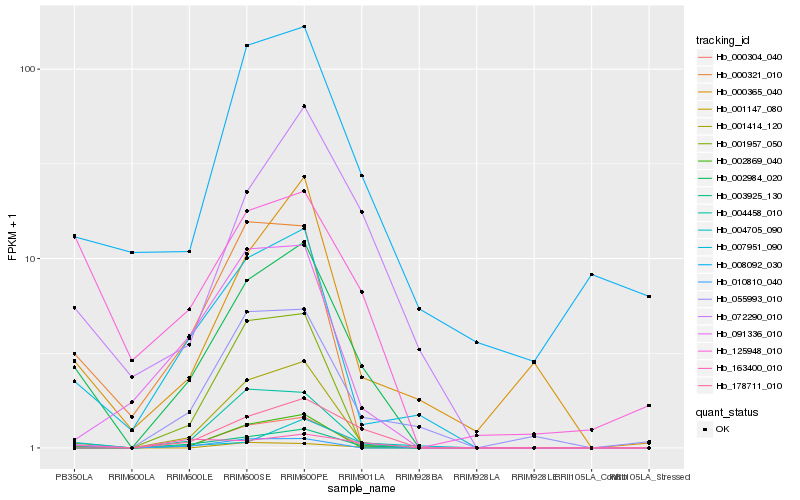
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002984_020 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_J02661 [Eucalyptus grandis] |
| 2 |
Hb_007951_090 |
0.1993352473 |
- |
- |
hypothetical protein POPTR_0003s06080g [Populus trichocarpa] |
| 3 |
Hb_003925_130 |
0.2066114496 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 4 |
Hb_072290_010 |
0.2088044141 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 5 |
Hb_010810_040 |
0.2203104106 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 6 |
Hb_000304_040 |
0.2210453611 |
- |
- |
PREDICTED: nicotianamine synthase-like [Jatropha curcas] |
| 7 |
Hb_178711_010 |
0.2215177496 |
- |
- |
hypothetical protein PHAVU_003G094000g [Phaseolus vulgaris] |
| 8 |
Hb_008092_030 |
0.2337305043 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 9 |
Hb_001147_080 |
0.2337622873 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001414_120 |
0.235297363 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000365_040 |
0.2365499229 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 12 |
Hb_000321_010 |
0.2373582215 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_125948_010 |
0.2378890515 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 14 |
Hb_163400_010 |
0.2446487483 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 15 |
Hb_002869_040 |
0.2459213204 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 16 |
Hb_004705_090 |
0.2471026384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001957_050 |
0.2541082317 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 18 |
Hb_055993_010 |
0.2557737136 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_004458_010 |
0.2583374284 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 20 |
Hb_091336_010 |
0.2594502519 |
- |
- |
- |