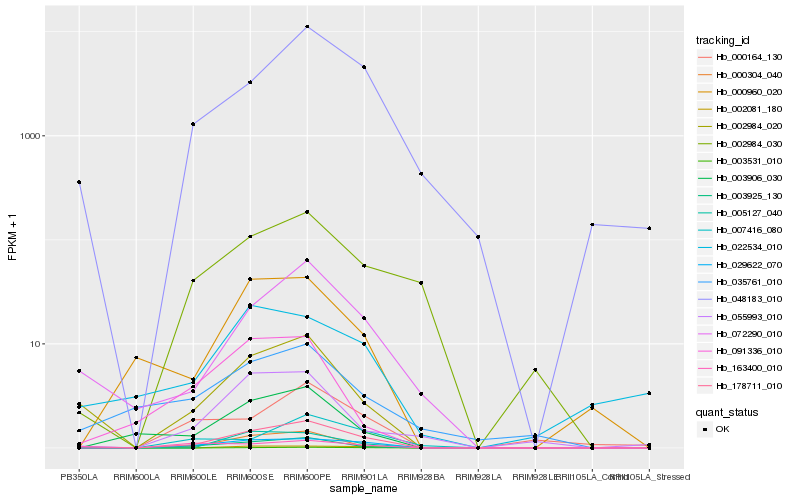
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029622_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_178711_010 |
0.1352787487 |
- |
- |
hypothetical protein PHAVU_003G094000g [Phaseolus vulgaris] |
| 3 |
Hb_002081_180 |
0.2239817631 |
- |
- |
hypothetical protein PRUPE_ppa006101mg [Prunus persica] |
| 4 |
Hb_003925_130 |
0.2247566898 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_003531_010 |
0.2455242322 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 6 |
Hb_048183_010 |
0.245990027 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_002984_030 |
0.25239429 |
- |
- |
- |
| 8 |
Hb_000960_020 |
0.2563611994 |
- |
- |
- |
| 9 |
Hb_000164_130 |
0.2571571978 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 10 |
Hb_007416_080 |
0.2602161607 |
- |
- |
- |
| 11 |
Hb_003906_030 |
0.2661139164 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_002984_020 |
0.2721106181 |
- |
- |
hypothetical protein EUGRSUZ_J02661 [Eucalyptus grandis] |
| 13 |
Hb_163400_010 |
0.2735454128 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 14 |
Hb_005127_040 |
0.2763406566 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 15 |
Hb_000304_040 |
0.2781101431 |
- |
- |
PREDICTED: nicotianamine synthase-like [Jatropha curcas] |
| 16 |
Hb_022534_010 |
0.2866849655 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 17 |
Hb_072290_010 |
0.2896772688 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 18 |
Hb_055993_010 |
0.2901542818 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_091336_010 |
0.2950397296 |
- |
- |
- |
| 20 |
Hb_035761_010 |
0.2958950016 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |