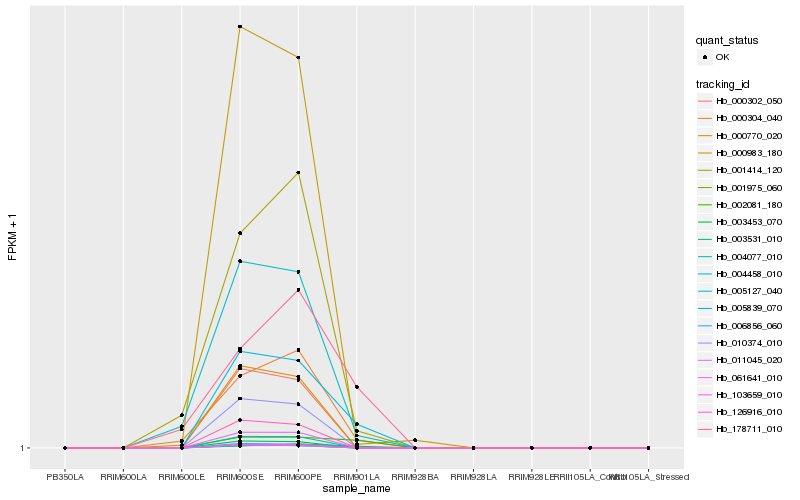
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005127_040 |
0.0 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 2 |
Hb_002081_180 |
0.1809130611 |
- |
- |
hypothetical protein PRUPE_ppa006101mg [Prunus persica] |
| 3 |
Hb_000304_040 |
0.1906932091 |
- |
- |
PREDICTED: nicotianamine synthase-like [Jatropha curcas] |
| 4 |
Hb_004458_010 |
0.1993258973 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 5 |
Hb_003531_010 |
0.2158248197 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 6 |
Hb_178711_010 |
0.2198111664 |
- |
- |
hypothetical protein PHAVU_003G094000g [Phaseolus vulgaris] |
| 7 |
Hb_000983_180 |
0.2217000751 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_001414_120 |
0.2273315983 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_005839_070 |
0.2290799034 |
- |
- |
hypothetical protein POPTR_0013s08160g [Populus trichocarpa] |
| 10 |
Hb_003453_070 |
0.2290824662 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 11 |
Hb_010374_010 |
0.2290843621 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_000770_020 |
0.2292762475 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 13 |
Hb_011045_020 |
0.2293249608 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 14 |
Hb_000302_050 |
0.2293796523 |
- |
- |
- |
| 15 |
Hb_126916_010 |
0.2294117113 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110, partial [Jatropha curcas] |
| 16 |
Hb_001975_060 |
0.2294622936 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 17 |
Hb_006856_060 |
0.2295998982 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 18 |
Hb_103659_010 |
0.2299924206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004077_010 |
0.23006921 |
- |
- |
retinoblastoma-binding protein, putative [Ricinus communis] |
| 20 |
Hb_061641_010 |
0.2303237544 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |