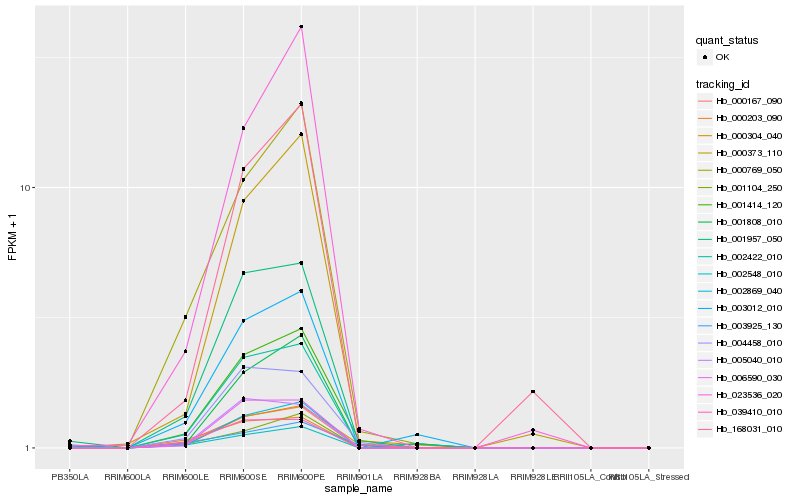
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001414_120 |
0.0 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000304_040 |
0.0475916291 |
- |
- |
PREDICTED: nicotianamine synthase-like [Jatropha curcas] |
| 3 |
Hb_004458_010 |
0.0951306995 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 4 |
Hb_000769_050 |
0.1072186761 |
- |
- |
hypothetical protein CISIN_1g030637mg [Citrus sinensis] |
| 5 |
Hb_001957_050 |
0.1072323024 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000167_090 |
0.113397013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002548_010 |
0.1216071954 |
- |
- |
hypothetical protein EUGRSUZ_L00530 [Eucalyptus grandis] |
| 8 |
Hb_000373_110 |
0.12963462 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 9 |
Hb_003925_130 |
0.1339512174 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 10 |
Hb_002422_010 |
0.1351886961 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 11 |
Hb_001104_250 |
0.1380946877 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 12 |
Hb_006590_030 |
0.1405391342 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_002869_040 |
0.1410414605 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 14 |
Hb_039410_010 |
0.1419436778 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 15 |
Hb_023536_020 |
0.1426433606 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 16 |
Hb_005040_010 |
0.1533656961 |
- |
- |
hypothetical protein RCOM_1110360 [Ricinus communis] |
| 17 |
Hb_001808_010 |
0.1538220979 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 18 |
Hb_168031_010 |
0.1541383233 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 19 |
Hb_003012_010 |
0.1557721313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000203_090 |
0.1589140397 |
- |
- |
hypothetical protein POPTR_0013s04080g [Populus trichocarpa] |