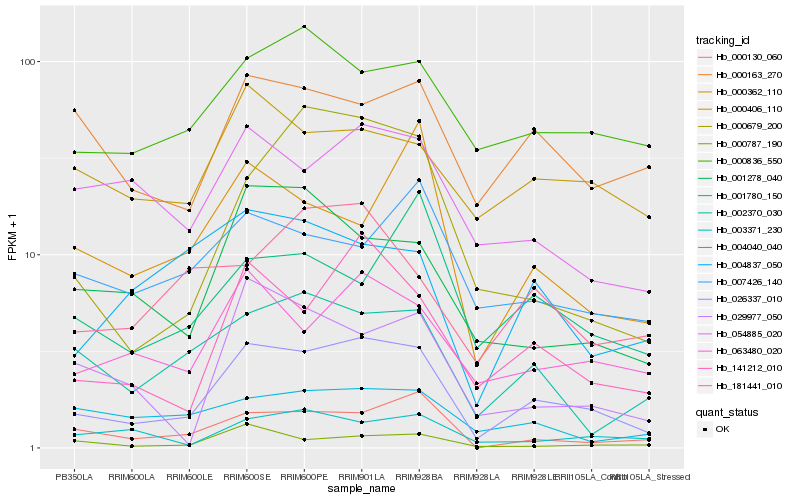
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026337_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 2 |
Hb_141212_010 |
0.1550854378 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_063480_020 |
0.1742460987 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000130_060 |
0.182280503 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 5 |
Hb_002370_030 |
0.1918840078 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 6 |
Hb_000679_200 |
0.1939962791 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 7 |
Hb_001278_040 |
0.1947625 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000836_550 |
0.1964858627 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 9 |
Hb_054885_020 |
0.2004304146 |
- |
- |
- |
| 10 |
Hb_000787_190 |
0.2049609924 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 15-like [Jatropha curcas] |
| 11 |
Hb_000163_270 |
0.2064014124 |
- |
- |
PREDICTED: uncharacterized protein LOC105642512 [Jatropha curcas] |
| 12 |
Hb_000406_110 |
0.2084701794 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 13 |
Hb_003371_230 |
0.2085940979 |
- |
- |
lactase-like b precursor [Scleropages formosus] |
| 14 |
Hb_029977_050 |
0.2100455192 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_181441_010 |
0.2120905756 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 16 |
Hb_004040_040 |
0.2129846634 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 17 |
Hb_004837_050 |
0.2132947237 |
- |
- |
- |
| 18 |
Hb_007426_140 |
0.2161063634 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_001780_150 |
0.2162272693 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 20 |
Hb_000362_110 |
0.2172389858 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |