| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
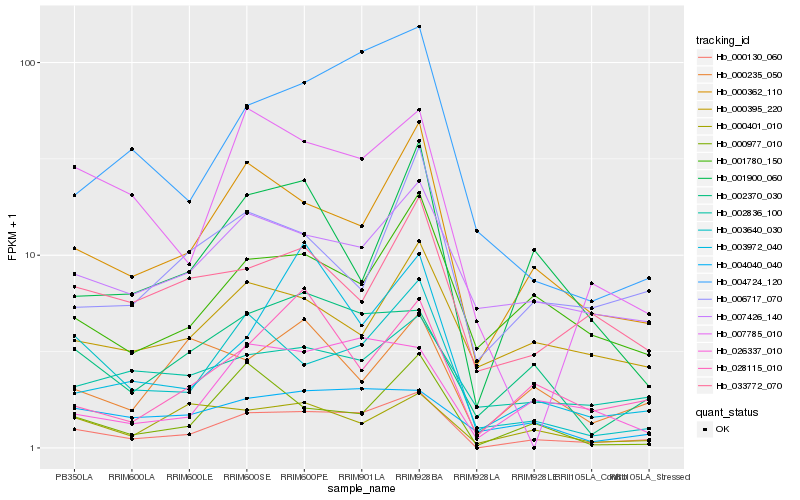
Hb_000130_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 2 |
Hb_000362_110 |
0.1447185705 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 3 |
Hb_002370_030 |
0.1822076442 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_026337_010 |
0.182280503 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 5 |
Hb_003640_030 |
0.1838460458 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004724_120 |
0.1881678861 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 7 |
Hb_007426_140 |
0.1924164592 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_001780_150 |
0.1927193694 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 9 |
Hb_001900_060 |
0.2025060345 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 10 |
Hb_028115_010 |
0.2032877289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002836_100 |
0.2058885371 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004040_040 |
0.2062067357 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 13 |
Hb_000977_010 |
0.2067500056 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_033772_070 |
0.207103422 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 15 |
Hb_007785_010 |
0.2110347554 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 16 |
Hb_000401_010 |
0.2136683296 |
- |
- |
SAB, putative [Ricinus communis] |
| 17 |
Hb_006717_070 |
0.2139793372 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 18 |
Hb_000395_220 |
0.2147714749 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 19 |
Hb_003972_040 |
0.2196779692 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 20 |
Hb_000235_050 |
0.2219719082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |