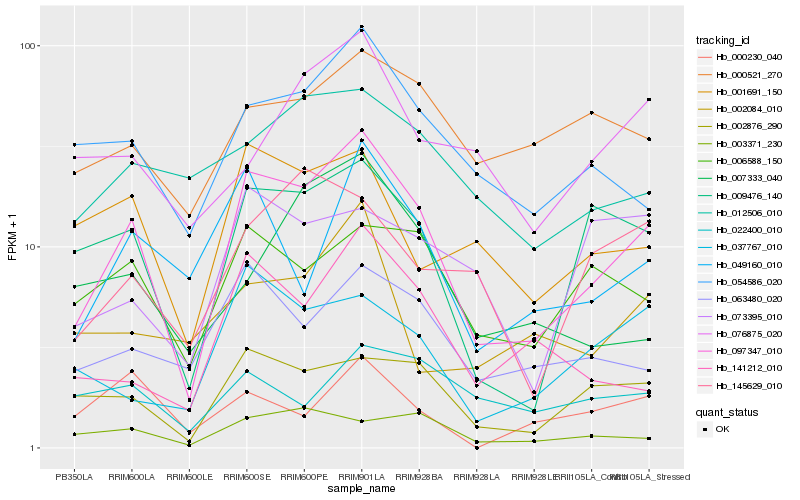
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097347_010 |
0.0 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |
| 2 |
Hb_009476_140 |
0.1799789342 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0838610 [Ricinus communis] |
| 3 |
Hb_002876_290 |
0.18261818 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 4 |
Hb_054586_020 |
0.1950329938 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 5 |
Hb_063480_020 |
0.2030258972 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_049160_010 |
0.2035715639 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_006588_150 |
0.2038214893 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 8 |
Hb_001691_150 |
0.2112209016 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Jatropha curcas] |
| 9 |
Hb_141212_010 |
0.2166159138 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_037767_010 |
0.2197294697 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_003371_230 |
0.2206039481 |
- |
- |
lactase-like b precursor [Scleropages formosus] |
| 12 |
Hb_012506_010 |
0.2238943046 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 13 |
Hb_000521_270 |
0.2273453614 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 14 |
Hb_145629_010 |
0.2323533558 |
- |
- |
- |
| 15 |
Hb_007333_040 |
0.2329031268 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 16 |
Hb_076875_020 |
0.2359965085 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 17 |
Hb_022400_010 |
0.2393641841 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 18 |
Hb_002084_010 |
0.2397140688 |
- |
- |
hypothetical protein VITISV_017990 [Vitis vinifera] |
| 19 |
Hb_073395_010 |
0.2397827839 |
- |
- |
- |
| 20 |
Hb_000230_040 |
0.2401694743 |
- |
- |
PREDICTED: protein MON2 homolog isoform X2 [Vitis vinifera] |