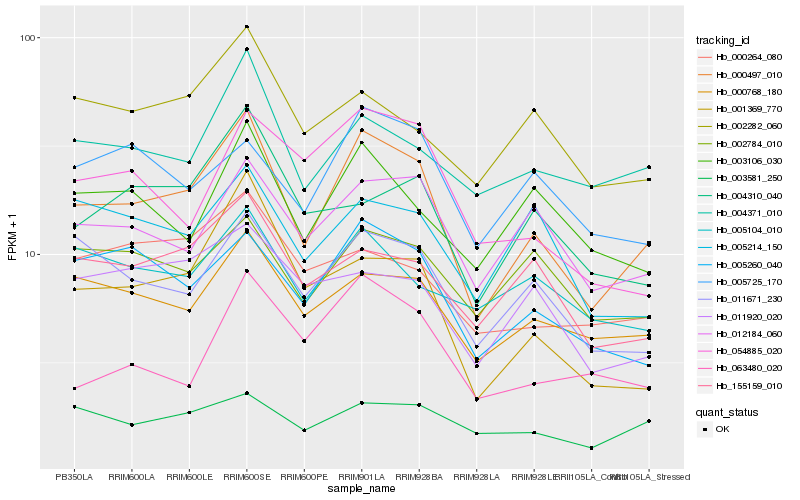
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_010 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR2 [Jatropha curcas] |
| 2 |
Hb_011671_230 |
0.1294401397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000768_180 |
0.1299911109 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 4 |
Hb_005260_040 |
0.1311361963 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012184_060 |
0.1318393633 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_155159_010 |
0.1409440666 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 7 |
Hb_001369_770 |
0.1421851212 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 8 |
Hb_004371_010 |
0.143703071 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_003106_030 |
0.1461937148 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 10 |
Hb_004310_040 |
0.1486330897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002282_060 |
0.1498952633 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_002784_010 |
0.1509890434 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 13 |
Hb_005725_170 |
0.1518071485 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 14 |
Hb_003581_250 |
0.1524371641 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 15 |
Hb_000264_080 |
0.1525396192 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 16 |
Hb_005214_150 |
0.1531355055 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 17 |
Hb_011920_020 |
0.1532449576 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 18 |
Hb_005104_010 |
0.1536485661 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 19 |
Hb_054885_020 |
0.1546872008 |
- |
- |
- |
| 20 |
Hb_063480_020 |
0.1553551488 |
- |
- |
unnamed protein product [Vitis vinifera] |