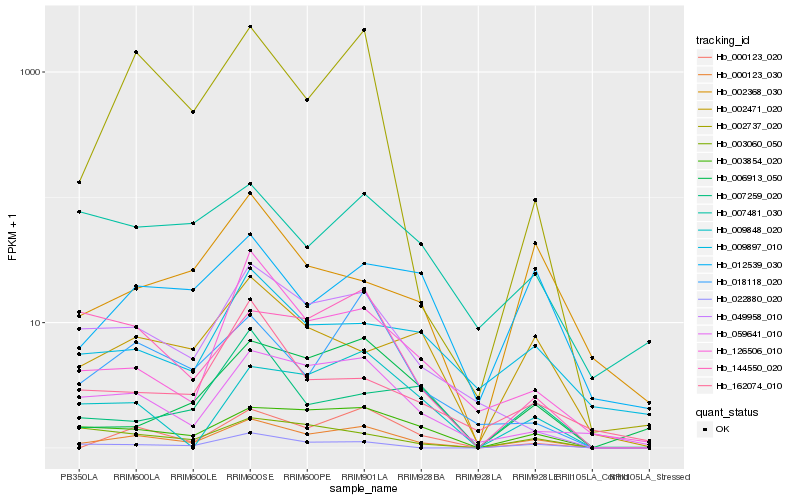
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_003854_020 |
0.1760284858 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_000123_020 |
0.1983674637 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 4 |
Hb_059641_010 |
0.2021822978 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_022880_020 |
0.2034710113 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 6 |
Hb_049958_010 |
0.2166354474 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_002471_020 |
0.2232329132 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 8 |
Hb_126506_010 |
0.2298223236 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_003060_050 |
0.2303934065 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_006913_050 |
0.2306349022 |
- |
- |
- |
| 11 |
Hb_012539_030 |
0.2307236344 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 12 |
Hb_009897_010 |
0.2314637611 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_009848_020 |
0.2333858502 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 14 |
Hb_002737_020 |
0.2342644563 |
- |
- |
metallothionein [Bruguiera gymnorhiza] |
| 15 |
Hb_018118_020 |
0.2373831273 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_144550_020 |
0.2510247343 |
- |
- |
- |
| 17 |
Hb_002368_030 |
0.2522856366 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 18 |
Hb_007259_020 |
0.2557148967 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 19 |
Hb_007481_030 |
0.2566560096 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 20 |
Hb_162074_010 |
0.2573964447 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |