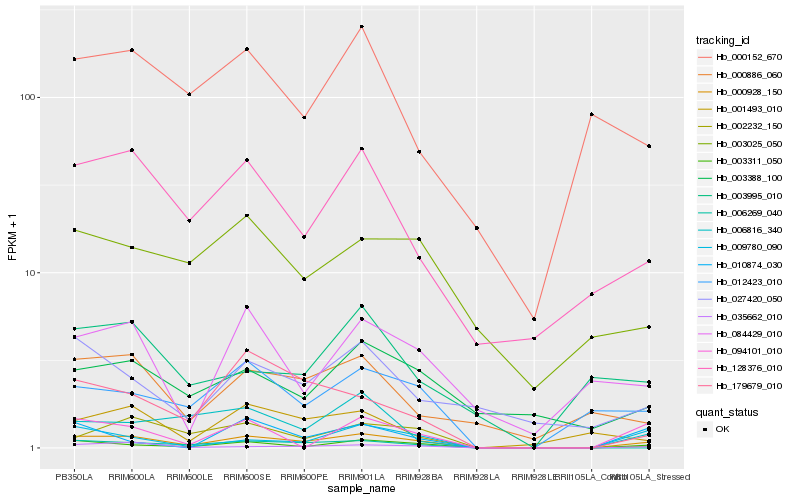
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003311_050 |
0.1517329161 |
- |
- |
PREDICTED: two-pore potassium channel 1-like [Jatropha curcas] |
| 3 |
Hb_035662_010 |
0.1852505734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009780_090 |
0.210017665 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 5 |
Hb_128376_010 |
0.2116807705 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_012423_010 |
0.2166380506 |
- |
- |
- |
| 7 |
Hb_006269_040 |
0.228939764 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 8 |
Hb_179679_010 |
0.2311207028 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_002232_150 |
0.2393176553 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 10 |
Hb_006816_340 |
0.2416677635 |
- |
- |
- |
| 11 |
Hb_094101_010 |
0.244978284 |
- |
- |
hypothetical protein JCGZ_18414 [Jatropha curcas] |
| 12 |
Hb_003388_100 |
0.2482631959 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 13 |
Hb_027420_050 |
0.2504544075 |
- |
- |
- |
| 14 |
Hb_000886_060 |
0.2505743807 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK10 isoform X2 [Vitis vinifera] |
| 15 |
Hb_084429_010 |
0.2525793394 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 16 |
Hb_001493_010 |
0.2539017738 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 17 |
Hb_003025_050 |
0.2551022668 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 18 |
Hb_010874_030 |
0.2554116927 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 19 |
Hb_000152_670 |
0.2563295753 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 20 |
Hb_003995_010 |
0.2565452913 |
- |
- |
- |