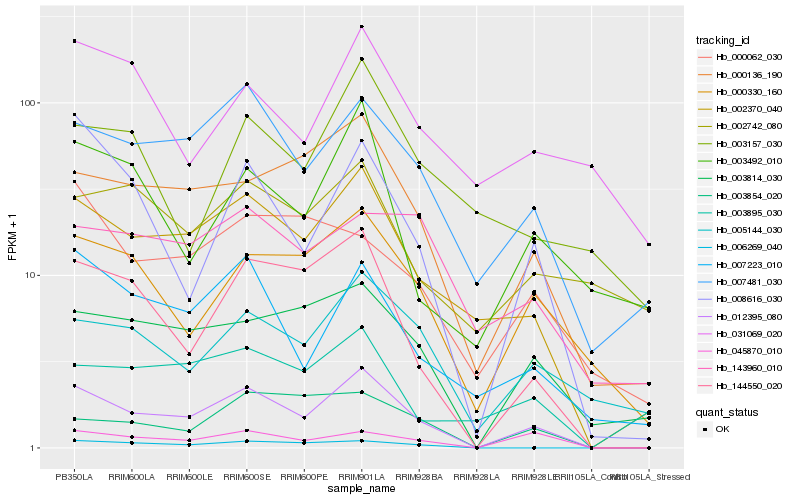
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_080 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 2 |
Hb_002370_040 |
0.145607366 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 3 |
Hb_144550_020 |
0.1573619249 |
- |
- |
- |
| 4 |
Hb_007481_030 |
0.1720440284 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 5 |
Hb_008616_030 |
0.1763825408 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000136_190 |
0.1793276848 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_045870_010 |
0.1817268223 |
- |
- |
- |
| 8 |
Hb_007223_010 |
0.1874456802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000330_160 |
0.1894698943 |
- |
- |
hypothetical protein JCGZ_04462 [Jatropha curcas] |
| 10 |
Hb_003814_030 |
0.1912147416 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_005144_030 |
0.1970683105 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 12 |
Hb_006269_040 |
0.204103597 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 13 |
Hb_003854_020 |
0.2055625811 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_003492_010 |
0.2185126604 |
- |
- |
DREPP plasma membrane polypeptide family protein [Populus trichocarpa] |
| 15 |
Hb_003895_030 |
0.2286668689 |
- |
- |
- |
| 16 |
Hb_000062_030 |
0.236099942 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 17 |
Hb_143960_010 |
0.2388870807 |
- |
- |
- |
| 18 |
Hb_003157_030 |
0.2401307656 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 19 |
Hb_002742_080 |
0.2406997792 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_031069_020 |
0.241884161 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |