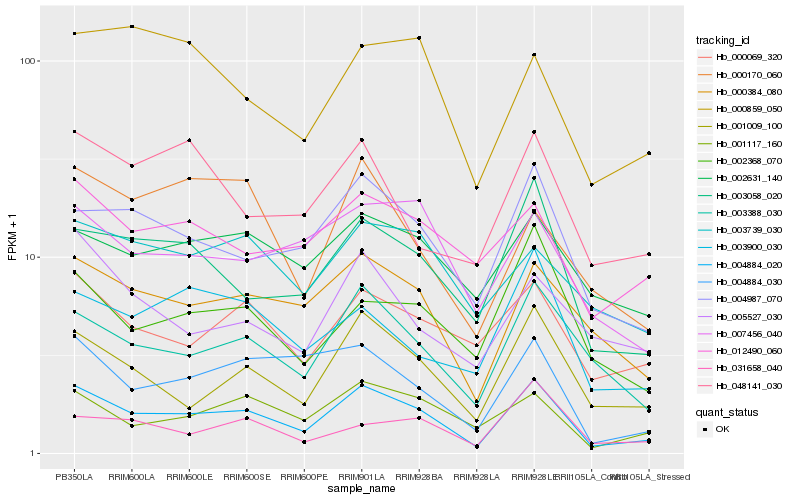
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004884_020 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001009_100 |
0.1277105494 |
- |
- |
- |
| 3 |
Hb_003388_030 |
0.1352523669 |
- |
- |
PREDICTED: uncharacterized protein LOC105649746 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003058_020 |
0.1359803138 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_004987_070 |
0.1359837274 |
- |
- |
PREDICTED: heat shock 70 kDa protein 17-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000384_080 |
0.1386404086 |
- |
- |
PREDICTED: RING-H2 finger protein ATL7-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_001117_160 |
0.1508057869 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 8 |
Hb_004884_030 |
0.1521273973 |
- |
- |
PREDICTED: ammonium transporter 3 member 3 [Vitis vinifera] |
| 9 |
Hb_000069_320 |
0.1585386178 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 10 |
Hb_002368_070 |
0.1613099065 |
- |
- |
PREDICTED: uncharacterized protein LOC105634087 [Jatropha curcas] |
| 11 |
Hb_003900_030 |
0.1666650597 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007456_040 |
0.1684428226 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 13 |
Hb_002631_140 |
0.1728219656 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 14 |
Hb_031658_040 |
0.1732608186 |
- |
- |
- |
| 15 |
Hb_012490_060 |
0.1735788578 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 16 |
Hb_048141_030 |
0.1744079077 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g59040-like [Prunus mume] |
| 17 |
Hb_005527_030 |
0.1760879698 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g42630 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003739_030 |
0.1768620648 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 19 |
Hb_000859_050 |
0.1774116204 |
- |
- |
hypothetical protein JCGZ_17054 [Jatropha curcas] |
| 20 |
Hb_000170_060 |
0.1778126964 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |