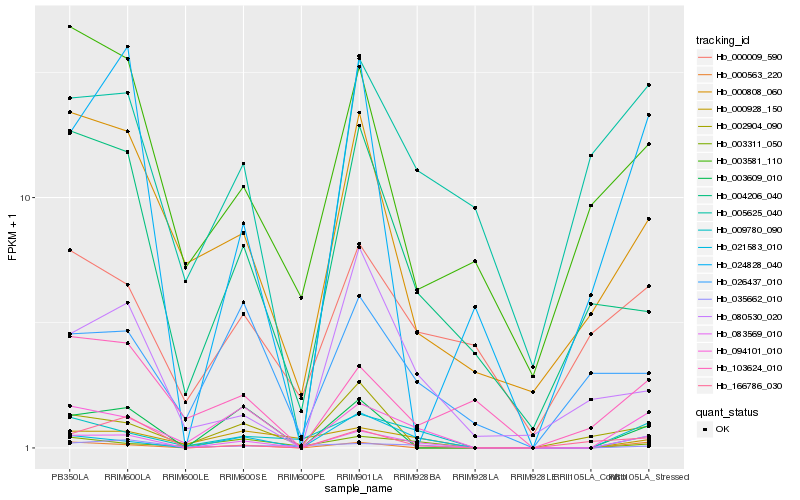
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083569_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_094101_010 |
0.160659514 |
- |
- |
hypothetical protein JCGZ_18414 [Jatropha curcas] |
| 3 |
Hb_021583_010 |
0.2134417606 |
- |
- |
- |
| 4 |
Hb_000563_220 |
0.222215683 |
- |
- |
hypothetical protein VITISV_004111 [Vitis vinifera] |
| 5 |
Hb_009780_090 |
0.2229930951 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 6 |
Hb_003609_010 |
0.2491117187 |
- |
- |
- |
| 7 |
Hb_003311_050 |
0.2793431731 |
- |
- |
PREDICTED: two-pore potassium channel 1-like [Jatropha curcas] |
| 8 |
Hb_024828_040 |
0.281524069 |
- |
- |
- |
| 9 |
Hb_000808_060 |
0.2868300737 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 10 |
Hb_004206_040 |
0.2904525708 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000928_150 |
0.2959059239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005625_040 |
0.2967623925 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 13 |
Hb_080530_020 |
0.3082125273 |
- |
- |
- |
| 14 |
Hb_166786_030 |
0.3108204854 |
- |
- |
PREDICTED: uncharacterized protein LOC105775425 [Gossypium raimondii] |
| 15 |
Hb_000009_590 |
0.3113779547 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_103624_010 |
0.3124205027 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 17 |
Hb_035662_010 |
0.3137615283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_026437_010 |
0.3149903883 |
- |
- |
- |
| 19 |
Hb_002904_090 |
0.3154598954 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420-like [Vitis vinifera] |
| 20 |
Hb_003581_110 |
0.3171121314 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |