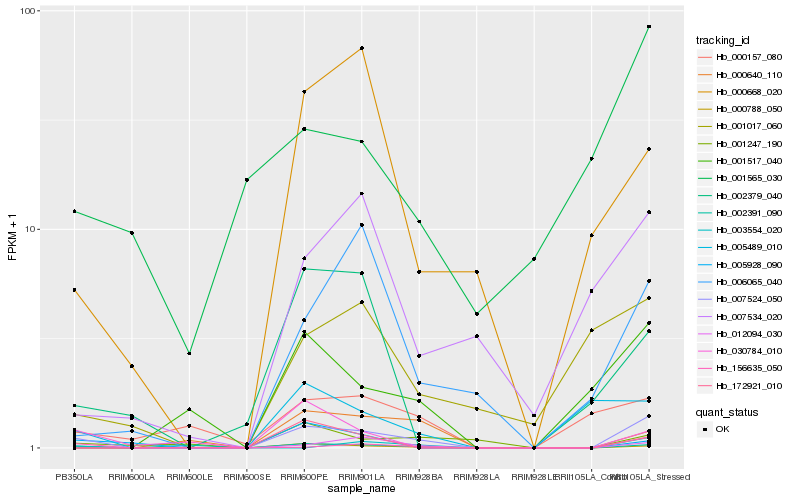
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007524_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 2 |
Hb_000788_050 |
0.1979434387 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 3 |
Hb_156635_050 |
0.3200855832 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 4 |
Hb_002391_090 |
0.3248718933 |
- |
- |
PREDICTED: IST1-like protein [Jatropha curcas] |
| 5 |
Hb_012094_030 |
0.3654286932 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 6 |
Hb_005489_010 |
0.3662996901 |
- |
- |
hypothetical protein POPTR_0011s11420g [Populus trichocarpa] |
| 7 |
Hb_003554_020 |
0.3742994865 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 8 |
Hb_001247_190 |
0.3784029582 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001517_040 |
0.3815828744 |
- |
- |
- |
| 10 |
Hb_000668_020 |
0.3851989411 |
- |
- |
- |
| 11 |
Hb_006065_040 |
0.3866094516 |
- |
- |
- |
| 12 |
Hb_030784_010 |
0.3870457629 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 13 |
Hb_000157_080 |
0.3890624326 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 14 |
Hb_002379_040 |
0.3960136474 |
- |
- |
- |
| 15 |
Hb_007534_020 |
0.396915163 |
- |
- |
- |
| 16 |
Hb_001017_060 |
0.3969829388 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 17 |
Hb_000640_110 |
0.398826174 |
- |
- |
- |
| 18 |
Hb_001565_030 |
0.4006948002 |
- |
- |
- |
| 19 |
Hb_005928_090 |
0.401166312 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_172921_010 |
0.4012707978 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |