| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
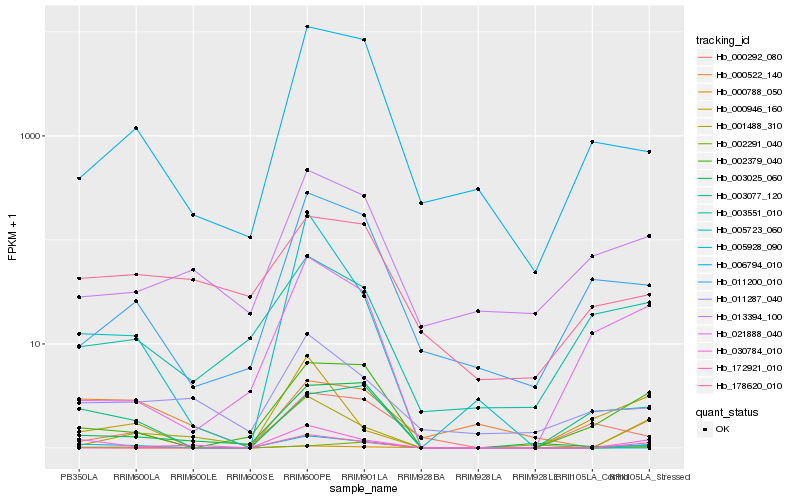
Hb_005928_090 |
0.0 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_030784_010 |
0.2160886099 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 3 |
Hb_002379_040 |
0.2719409541 |
- |
- |
- |
| 4 |
Hb_000788_050 |
0.296893942 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 5 |
Hb_003077_120 |
0.3067973517 |
- |
- |
- |
| 6 |
Hb_001488_310 |
0.3182620147 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_000522_140 |
0.3187834525 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 8 |
Hb_006794_010 |
0.3226858015 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 9 |
Hb_011200_010 |
0.3241323856 |
- |
- |
- |
| 10 |
Hb_021888_040 |
0.3361509699 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 11 |
Hb_172921_010 |
0.3404092565 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 12 |
Hb_003025_060 |
0.3420267643 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 13 |
Hb_011287_040 |
0.3437012831 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003551_010 |
0.3443513541 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 15 |
Hb_000946_160 |
0.3470252342 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 16 |
Hb_002291_040 |
0.3482763719 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 17 |
Hb_005723_060 |
0.3492466179 |
- |
- |
- |
| 18 |
Hb_013394_100 |
0.3524890534 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 19 |
Hb_178620_010 |
0.3532726821 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 20 |
Hb_000292_080 |
0.353900282 |
- |
- |
hypothetical protein CISIN_1g034193mg [Citrus sinensis] |