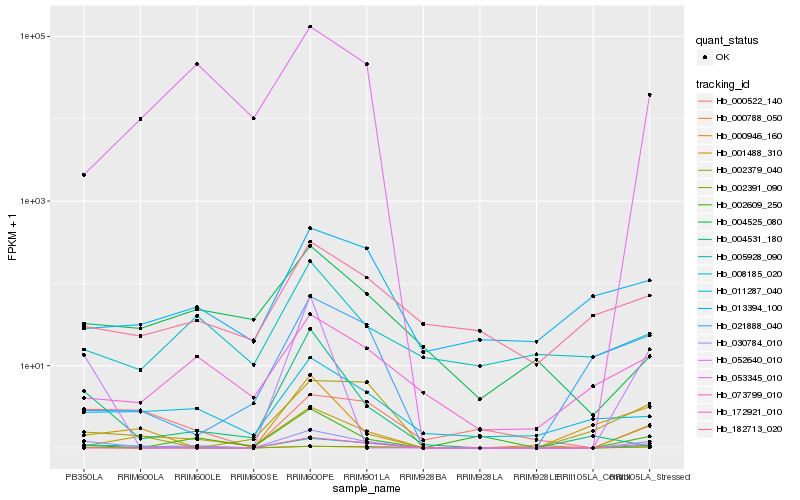
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030784_010 |
0.0 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 2 |
Hb_005928_090 |
0.2160886099 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_000788_050 |
0.2909976013 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 4 |
Hb_001488_310 |
0.2922381349 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_011287_040 |
0.3160500957 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002391_090 |
0.3290920305 |
- |
- |
PREDICTED: IST1-like protein [Jatropha curcas] |
| 7 |
Hb_073799_010 |
0.3351749272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_013394_100 |
0.3404379837 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 9 |
Hb_053345_010 |
0.3424927702 |
- |
- |
- |
| 10 |
Hb_182713_020 |
0.3435487082 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 11 |
Hb_002379_040 |
0.3461174229 |
- |
- |
- |
| 12 |
Hb_052640_010 |
0.3500488142 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004531_180 |
0.3525966635 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_000522_140 |
0.3526932502 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_004525_080 |
0.3528705929 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 16 |
Hb_021888_040 |
0.3533922818 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 17 |
Hb_172921_010 |
0.3536949868 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 18 |
Hb_000946_160 |
0.3545533112 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 19 |
Hb_008185_020 |
0.3590754743 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 20 |
Hb_002609_250 |
0.3591688794 |
- |
- |
PREDICTED: starch-binding domain-containing protein 1-like [Jatropha curcas] |