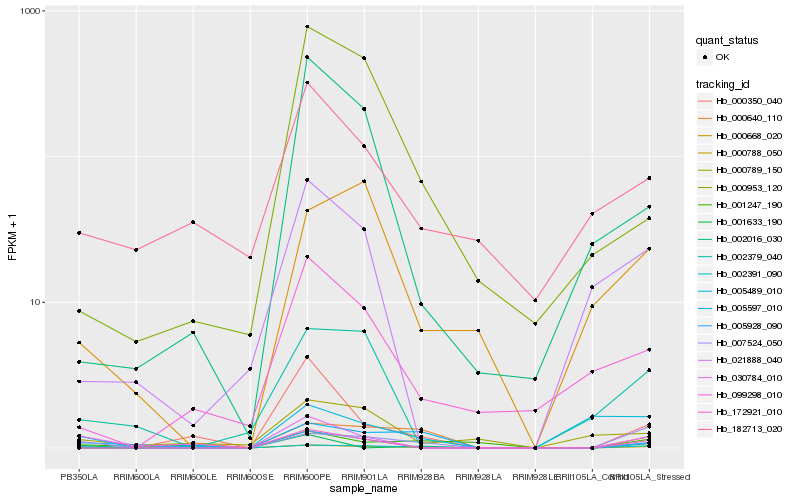
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000788_050 |
0.0 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 2 |
Hb_007524_050 |
0.1979434387 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 3 |
Hb_030784_010 |
0.2909976013 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 4 |
Hb_005928_090 |
0.296893942 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_172921_010 |
0.3128824988 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 6 |
Hb_002391_090 |
0.3129281685 |
- |
- |
PREDICTED: IST1-like protein [Jatropha curcas] |
| 7 |
Hb_002379_040 |
0.331313286 |
- |
- |
- |
| 8 |
Hb_001247_190 |
0.336368115 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000668_020 |
0.3408386413 |
- |
- |
- |
| 10 |
Hb_001633_190 |
0.3416911976 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 11 |
Hb_005489_010 |
0.3427188161 |
- |
- |
hypothetical protein POPTR_0011s11420g [Populus trichocarpa] |
| 12 |
Hb_000640_110 |
0.3449904551 |
- |
- |
- |
| 13 |
Hb_005597_010 |
0.3490484941 |
- |
- |
- |
| 14 |
Hb_099298_010 |
0.3494352409 |
- |
- |
- |
| 15 |
Hb_021888_040 |
0.3590364349 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 16 |
Hb_000350_040 |
0.3593151036 |
- |
- |
- |
| 17 |
Hb_002016_030 |
0.3605103706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000789_150 |
0.3606433473 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000953_120 |
0.3616467836 |
- |
- |
- |
| 20 |
Hb_182713_020 |
0.3629728149 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |