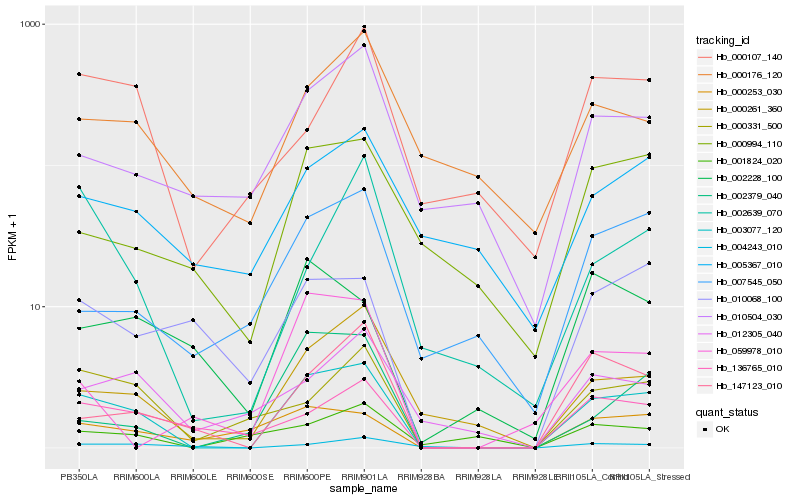
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003077_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_000261_360 |
0.2380117194 |
- |
- |
unknown [Lotus japonicus] |
| 3 |
Hb_000331_500 |
0.2437418369 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_147123_010 |
0.2480244629 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_136765_010 |
0.2521035215 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_004243_010 |
0.2557234273 |
- |
- |
hypothetical protein JCGZ_01580 [Jatropha curcas] |
| 7 |
Hb_000253_030 |
0.2607263731 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 8 |
Hb_002379_040 |
0.2671512228 |
- |
- |
- |
| 9 |
Hb_010068_100 |
0.2728736535 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007545_050 |
0.2763849364 |
- |
- |
- |
| 11 |
Hb_000994_110 |
0.2773346678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000107_140 |
0.277847564 |
- |
- |
- |
| 13 |
Hb_002639_070 |
0.2800805939 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 14 |
Hb_012305_040 |
0.2808962604 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A-like [Jatropha curcas] |
| 15 |
Hb_010504_030 |
0.282248077 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 16 |
Hb_002228_100 |
0.285023062 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_059978_010 |
0.2908179898 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 18 |
Hb_001824_020 |
0.2915995225 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 19 |
Hb_005367_010 |
0.2928452954 |
- |
- |
- |
| 20 |
Hb_000176_120 |
0.2930199609 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |