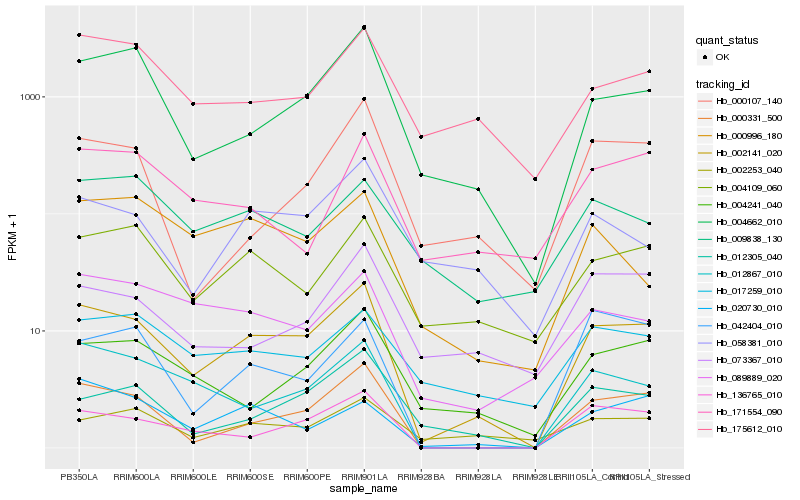
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002141_020 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 2 |
Hb_000331_500 |
0.138158489 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_136765_010 |
0.156156616 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004662_010 |
0.1654161152 |
- |
- |
Nascent polypeptide-associated complex subunit beta [Medicago truncatula] |
| 5 |
Hb_012867_010 |
0.1698026535 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 6 |
Hb_004109_060 |
0.1807706463 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 7 |
Hb_004241_040 |
0.1832708882 |
- |
- |
- |
| 8 |
Hb_012305_040 |
0.1856084176 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A-like [Jatropha curcas] |
| 9 |
Hb_020730_010 |
0.191180315 |
- |
- |
hypothetical protein CICLE_v10010243mg, partial [Citrus clementina] |
| 10 |
Hb_089889_020 |
0.192277165 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 11 |
Hb_175612_010 |
0.1924915564 |
transcription factor |
TF Family: MBF1 |
orf [Ricinus communis] |
| 12 |
Hb_002253_040 |
0.1939599419 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 13 |
Hb_017259_010 |
0.1956997351 |
- |
- |
- |
| 14 |
Hb_058381_010 |
0.1973136292 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 15 |
Hb_000107_140 |
0.2048666631 |
- |
- |
- |
| 16 |
Hb_000996_180 |
0.2048870691 |
- |
- |
PREDICTED: tetraspanin-10 [Jatropha curcas] |
| 17 |
Hb_042404_010 |
0.2082942969 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 18 |
Hb_073367_010 |
0.2088474756 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 19 |
Hb_009838_130 |
0.2100557203 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 20 |
Hb_171554_090 |
0.2104244885 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |