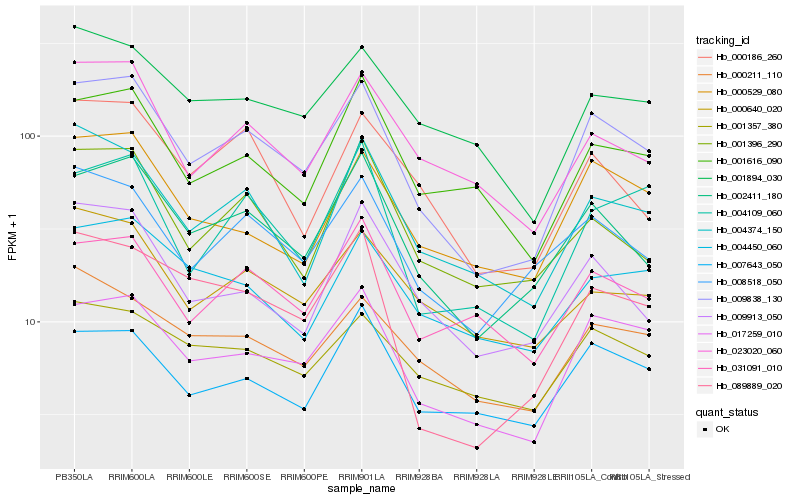
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_130 |
0.0 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 2 |
Hb_017259_010 |
0.0713812568 |
- |
- |
- |
| 3 |
Hb_002411_180 |
0.0774280244 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008518_050 |
0.1021615183 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 5 |
Hb_004374_150 |
0.1055548733 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000529_080 |
0.1080761574 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |
| 7 |
Hb_007643_050 |
0.1083381284 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000211_110 |
0.1098365333 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_023020_060 |
0.1111221187 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 10 |
Hb_000186_260 |
0.1119027343 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 11 |
Hb_004109_060 |
0.1125738769 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 12 |
Hb_009913_050 |
0.1139739493 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 13 |
Hb_001616_090 |
0.1146343087 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_089889_020 |
0.1151554669 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 15 |
Hb_001357_380 |
0.1154381428 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_031091_010 |
0.1157063651 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 17 |
Hb_001894_030 |
0.1164618658 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 18 |
Hb_001396_290 |
0.1168480344 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 19 |
Hb_004450_060 |
0.1190995079 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000640_020 |
0.1198291218 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |