| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
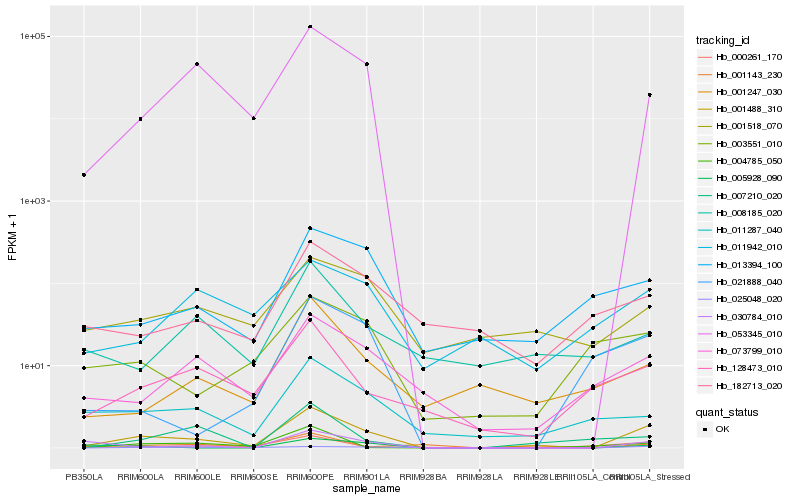
Hb_001488_310 |
0.0 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_053345_010 |
0.2536456579 |
- |
- |
- |
| 3 |
Hb_073799_010 |
0.2841754065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_030784_010 |
0.2922381349 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 5 |
Hb_128473_010 |
0.2931254024 |
- |
- |
- |
| 6 |
Hb_013394_100 |
0.2932337079 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 7 |
Hb_011287_040 |
0.2993718582 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001518_070 |
0.3067601013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001247_030 |
0.3107460602 |
- |
- |
- |
| 10 |
Hb_000261_170 |
0.3136453144 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_182713_020 |
0.314813171 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 12 |
Hb_003551_010 |
0.3152743179 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 13 |
Hb_004785_050 |
0.317357701 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 14 |
Hb_011942_010 |
0.3182010563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005928_090 |
0.3182620147 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_001143_230 |
0.3184705806 |
- |
- |
beta-galactosidase family protein [Populus trichocarpa] |
| 17 |
Hb_025048_020 |
0.3194788148 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_021888_040 |
0.3238927348 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 19 |
Hb_008185_020 |
0.324989194 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 20 |
Hb_007210_020 |
0.3267177504 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |