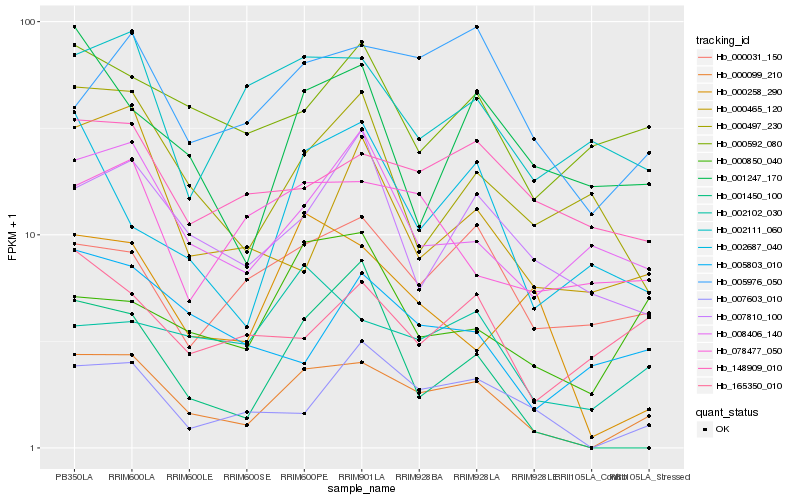
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 2 |
Hb_007603_010 |
0.1896488129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008406_140 |
0.2141402083 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 4 |
Hb_005976_050 |
0.2155561849 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 5 |
Hb_000031_150 |
0.2185990522 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_002102_030 |
0.2197545317 |
- |
- |
- |
| 7 |
Hb_007810_100 |
0.2222080359 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 8 |
Hb_000258_290 |
0.2227201489 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 9 |
Hb_002111_060 |
0.2258582073 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 10 |
Hb_002687_040 |
0.226101728 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 11 |
Hb_148909_010 |
0.2265403864 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 12 |
Hb_078477_050 |
0.2267228111 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000465_120 |
0.2300261657 |
- |
- |
PREDICTED: uncharacterized protein LOC105632294 [Jatropha curcas] |
| 14 |
Hb_165350_010 |
0.230087395 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 15 |
Hb_001247_170 |
0.230489012 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 16 |
Hb_000592_080 |
0.2305243178 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 17 |
Hb_001450_100 |
0.2324942733 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 18 |
Hb_005803_010 |
0.2327795848 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000850_040 |
0.2333242689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000497_230 |
0.2333849501 |
- |
- |
PREDICTED: probable protein S-acyltransferase 6 [Jatropha curcas] |