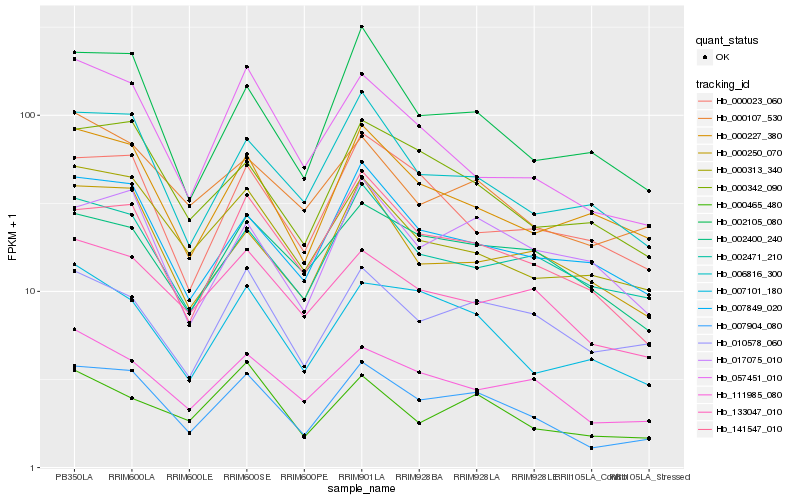
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 2 |
Hb_141547_010 |
0.1047470158 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 3 |
Hb_000342_090 |
0.1111356188 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_111985_080 |
0.1111933881 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 5 |
Hb_002105_080 |
0.1123744053 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 6 |
Hb_006816_300 |
0.1129094735 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 7 |
Hb_007849_020 |
0.1142782029 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 8 |
Hb_133047_010 |
0.1158070819 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_000313_340 |
0.1168904827 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 10 |
Hb_010578_060 |
0.1168983096 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 11 |
Hb_000227_380 |
0.1180273435 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 12 |
Hb_007101_180 |
0.1183639575 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 13 |
Hb_002471_210 |
0.1189211064 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 14 |
Hb_057451_010 |
0.1219771195 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 15 |
Hb_000023_060 |
0.1229485494 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 16 |
Hb_002400_240 |
0.1234772611 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 17 |
Hb_000107_530 |
0.1237577782 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000465_480 |
0.1241412481 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000250_070 |
0.1242826868 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase listerin [Jatropha curcas] |
| 20 |
Hb_017075_010 |
0.1248280668 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 1 [Jatropha curcas] |