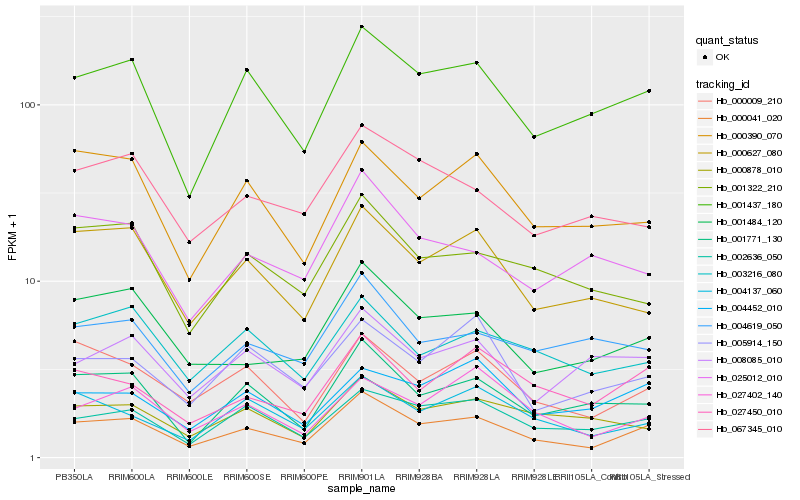
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s11640g [Populus trichocarpa] |
| 2 |
Hb_001437_180 |
0.1023559078 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 3 |
Hb_002636_050 |
0.1174274087 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g68930 [Jatropha curcas] |
| 4 |
Hb_001484_120 |
0.1184970664 |
- |
- |
hypothetical protein POPTR_2287s00200g, partial [Populus trichocarpa] |
| 5 |
Hb_001771_130 |
0.1193207003 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g27610 [Jatropha curcas] |
| 6 |
Hb_000009_210 |
0.1197132264 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01390 [Jatropha curcas] |
| 7 |
Hb_027450_010 |
0.1260805596 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Populus euphratica] |
| 8 |
Hb_000627_080 |
0.1292692737 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 9 |
Hb_004137_060 |
0.1304682811 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09040, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000390_070 |
0.1340074315 |
- |
- |
PREDICTED: uncharacterized protein LOC105632478 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003216_080 |
0.1363680335 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560-like [Jatropha curcas] |
| 12 |
Hb_001322_210 |
0.1384243852 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 13 |
Hb_027402_140 |
0.1388632986 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g47530 [Jatropha curcas] |
| 14 |
Hb_025012_010 |
0.1394062239 |
- |
- |
PREDICTED: importin-4 [Vitis vinifera] |
| 15 |
Hb_067345_010 |
0.1401568353 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 16 |
Hb_004619_050 |
0.1407136345 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_005914_150 |
0.1407286053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 18 |
Hb_004452_010 |
0.143269148 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 19 |
Hb_008085_010 |
0.1433401369 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g26680, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000878_010 |
0.1435119146 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g06400, mitochondrial [Jatropha curcas] |