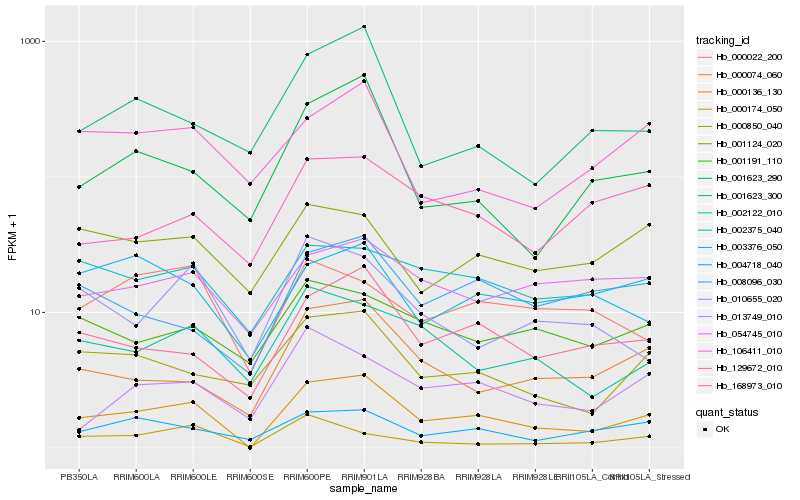
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000136_130 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 7a [Solanum lycopersicum] |
| 2 |
Hb_168973_010 |
0.1688845634 |
- |
- |
- |
| 3 |
Hb_000074_060 |
0.1827076761 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 4 |
Hb_000850_040 |
0.1881867246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004718_040 |
0.1896404893 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 6 |
Hb_002122_010 |
0.1910279531 |
- |
- |
- |
| 7 |
Hb_001623_290 |
0.1982596493 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_008096_030 |
0.2029364019 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 9 |
Hb_002375_040 |
0.2042048796 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 10 |
Hb_001623_300 |
0.2050400433 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 11 |
Hb_013749_010 |
0.2054529601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_054745_010 |
0.2072605022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001124_020 |
0.2088387336 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 14 |
Hb_003376_050 |
0.2091109494 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 15 |
Hb_001191_110 |
0.2092179766 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 16 |
Hb_106411_010 |
0.2111058538 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 17 |
Hb_129672_010 |
0.2112524357 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 18 |
Hb_010655_020 |
0.2117471397 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 19 |
Hb_000022_200 |
0.2145234632 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 20 |
Hb_000174_050 |
0.215643521 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |