| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
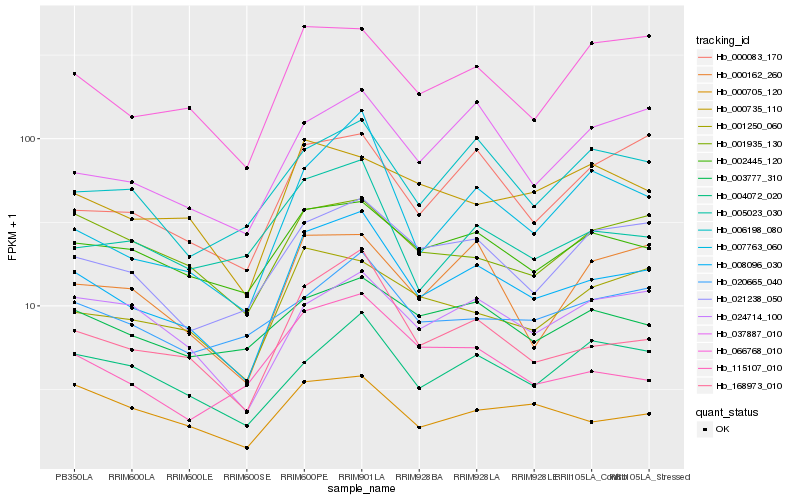
Hb_008096_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 2 |
Hb_168973_010 |
0.0798983935 |
- |
- |
- |
| 3 |
Hb_002445_120 |
0.1169534831 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 4 |
Hb_021238_050 |
0.1176141633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_066768_010 |
0.1176229302 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_001250_060 |
0.1238442923 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 7 |
Hb_007763_060 |
0.1273439045 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 8 |
Hb_004072_020 |
0.1273643952 |
- |
- |
PREDICTED: uncharacterized protein LOC105635274 [Jatropha curcas] |
| 9 |
Hb_000162_260 |
0.1285553247 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1 [Nelumbo nucifera] |
| 10 |
Hb_115107_010 |
0.1300161988 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 11 |
Hb_000705_120 |
0.130067009 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005023_030 |
0.1301084732 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 13 |
Hb_006198_080 |
0.130126829 |
- |
- |
hypothetical protein MIMGU_mgv1a013452mg [Erythranthe guttata] |
| 14 |
Hb_000083_170 |
0.1318041525 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 7-like [Populus euphratica] |
| 15 |
Hb_037887_010 |
0.1318585394 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 16 |
Hb_024714_100 |
0.1321646679 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001935_130 |
0.133664661 |
- |
- |
PREDICTED: uncharacterized protein LOC105643176 [Jatropha curcas] |
| 18 |
Hb_003777_310 |
0.1341414127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000735_110 |
0.1350172671 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 20 |
Hb_020665_040 |
0.1364097812 |
transcription factor |
TF Family: Jumonji |
PREDICTED: trichohyalin-like [Malus domestica] |