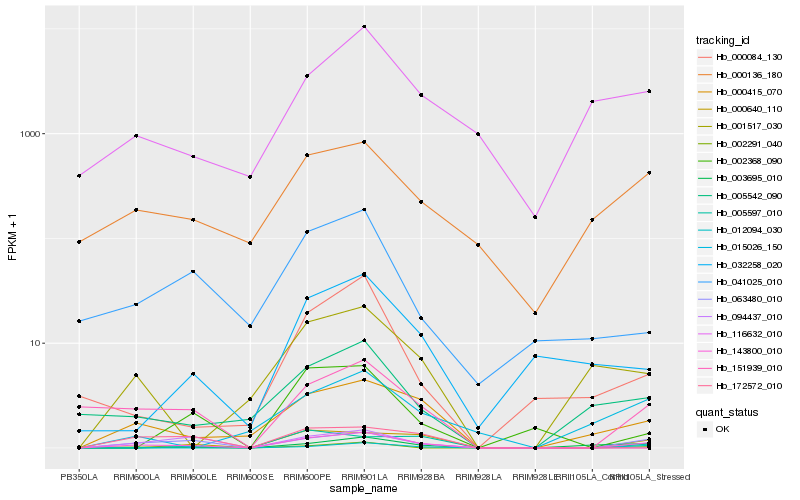
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143800_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s21410g, partial [Populus trichocarpa] |
| 2 |
Hb_063480_010 |
0.1685756863 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like isoform X2 [Pyrus x bretschneideri] |
| 3 |
Hb_000415_070 |
0.2241860318 |
- |
- |
- |
| 4 |
Hb_151939_010 |
0.2317932847 |
- |
- |
PREDICTED: far upstream element-binding protein 2 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000640_110 |
0.2569472588 |
- |
- |
- |
| 6 |
Hb_000136_180 |
0.2936905324 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 7 |
Hb_005542_090 |
0.2957498387 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 8 |
Hb_002291_040 |
0.3000702022 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 9 |
Hb_012094_030 |
0.3025438535 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 10 |
Hb_001517_030 |
0.3038403375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005597_010 |
0.3059569356 |
- |
- |
- |
| 12 |
Hb_002368_090 |
0.3079744554 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 13 |
Hb_003695_010 |
0.3086128806 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 14 |
Hb_116632_010 |
0.3112701415 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000084_130 |
0.3144031604 |
- |
- |
- |
| 16 |
Hb_041025_010 |
0.3170004675 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 17 |
Hb_172572_010 |
0.3175771976 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 18 |
Hb_032258_020 |
0.3185177382 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 19 |
Hb_015026_150 |
0.3219831704 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 20 |
Hb_094437_010 |
0.324719869 |
- |
- |
- |