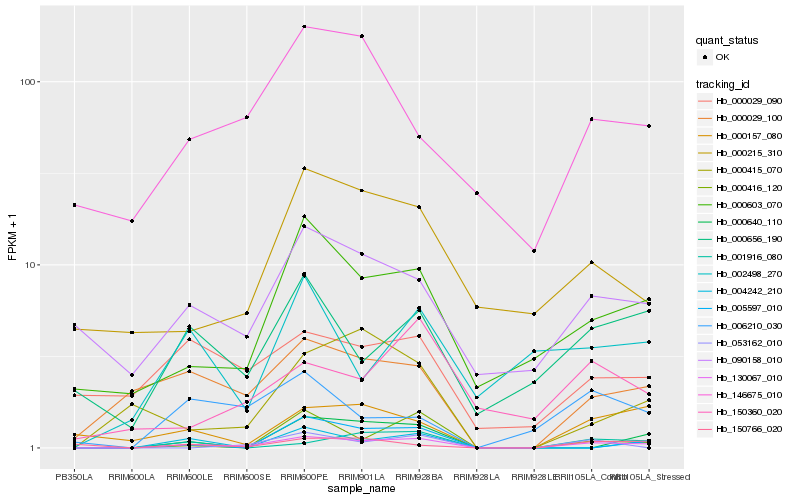
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130067_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000029_100 |
0.2573249194 |
- |
- |
PREDICTED: nicalin-1-like isoform X2 [Populus euphratica] |
| 3 |
Hb_000603_070 |
0.2593359137 |
- |
- |
- |
| 4 |
Hb_001916_080 |
0.2820620461 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 5 |
Hb_000157_080 |
0.2870686457 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 6 |
Hb_090158_010 |
0.2873653784 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 7 |
Hb_000656_190 |
0.2879917559 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |
| 8 |
Hb_000416_120 |
0.2942344328 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 9 |
Hb_006210_030 |
0.298710678 |
- |
- |
- |
| 10 |
Hb_000415_070 |
0.2997693679 |
- |
- |
- |
| 11 |
Hb_000215_310 |
0.3014811682 |
- |
- |
- |
| 12 |
Hb_150766_020 |
0.3033946712 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 13 |
Hb_000640_110 |
0.3049771765 |
- |
- |
- |
| 14 |
Hb_146675_010 |
0.3069640732 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 15 |
Hb_000029_090 |
0.3108208922 |
- |
- |
Nicalin precursor, putative [Ricinus communis] |
| 16 |
Hb_150360_020 |
0.3114274401 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 17 |
Hb_004242_210 |
0.3146898977 |
- |
- |
PREDICTED: uncharacterized protein LOC105633248 [Jatropha curcas] |
| 18 |
Hb_053162_010 |
0.3154927402 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 19 |
Hb_005597_010 |
0.3182938024 |
- |
- |
- |
| 20 |
Hb_002498_270 |
0.3217415306 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |