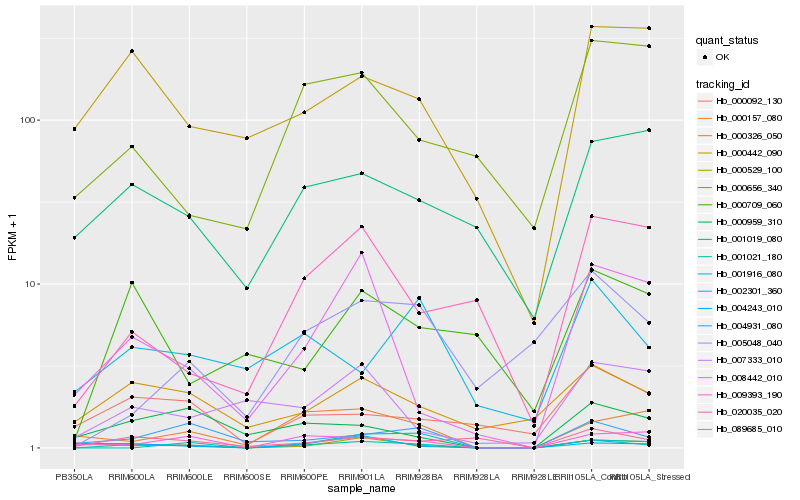
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_180 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 2 |
Hb_009393_190 |
0.2733658204 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 3 |
Hb_008442_010 |
0.2857679221 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 4 |
Hb_004931_080 |
0.2913532452 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Jatropha curcas] |
| 5 |
Hb_000326_050 |
0.2983893728 |
- |
- |
Phenylalanyl-tRNA synthetase alpha chain isoform 2 [Theobroma cacao] |
| 6 |
Hb_000157_080 |
0.3042491981 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 7 |
Hb_089685_010 |
0.3058002159 |
- |
- |
- |
| 8 |
Hb_000529_100 |
0.3095408528 |
- |
- |
hypothetical protein POPTR_0009s05980g [Populus trichocarpa] |
| 9 |
Hb_000959_310 |
0.3097977713 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 10 |
Hb_001916_080 |
0.310416415 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 11 |
Hb_000442_090 |
0.3137669078 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 12 |
Hb_020035_020 |
0.3163148148 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 13 |
Hb_000092_130 |
0.3174680615 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_005048_040 |
0.317843025 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 15 |
Hb_001019_080 |
0.3208258155 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 16 |
Hb_004243_010 |
0.3220160408 |
- |
- |
hypothetical protein JCGZ_01580 [Jatropha curcas] |
| 17 |
Hb_007333_010 |
0.3230359977 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000709_060 |
0.3280863332 |
- |
- |
PREDICTED: abscisic-aldehyde oxidase-like isoform X5 [Populus euphratica] |
| 19 |
Hb_000656_340 |
0.3281309429 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 20 |
Hb_002301_360 |
0.3321159853 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |