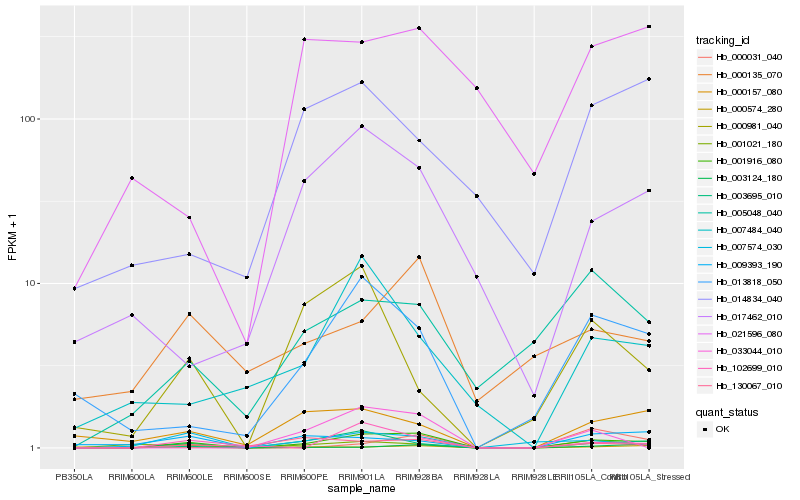
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001916_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 2 |
Hb_003695_010 |
0.2775759895 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 3 |
Hb_130067_010 |
0.2820620461 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_013818_050 |
0.2959689908 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001021_180 |
0.310416415 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 6 |
Hb_102699_010 |
0.3133991581 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR type disease resistance protein [Populus trichocarpa] |
| 7 |
Hb_007574_030 |
0.3204169285 |
- |
- |
hypothetical protein POPTR_0006s24510g [Populus trichocarpa] |
| 8 |
Hb_017462_010 |
0.3234932723 |
- |
- |
- |
| 9 |
Hb_000157_080 |
0.3239829011 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 10 |
Hb_005048_040 |
0.3345275438 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 11 |
Hb_007484_040 |
0.3361247534 |
- |
- |
- |
| 12 |
Hb_000135_070 |
0.3462030825 |
- |
- |
PREDICTED: inositol oxygenase 1-like [Jatropha curcas] |
| 13 |
Hb_009393_190 |
0.3511838032 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 14 |
Hb_033044_010 |
0.3537367489 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5-like isoform X3 [Gossypium raimondii] |
| 15 |
Hb_000031_040 |
0.3606012493 |
- |
- |
centromere protein O [Medicago truncatula] |
| 16 |
Hb_000981_040 |
0.363685347 |
- |
- |
- |
| 17 |
Hb_021596_080 |
0.3638880414 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 18 |
Hb_014834_040 |
0.365113747 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 19 |
Hb_000574_280 |
0.3685079611 |
- |
- |
hypothetical protein POPTR_0006s28780g [Populus trichocarpa] |
| 20 |
Hb_003124_180 |
0.3698471754 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |