| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
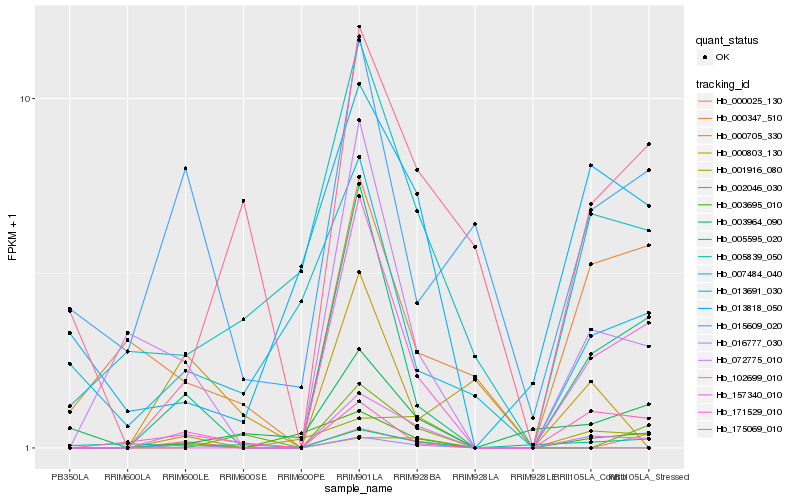
Hb_102699_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR type disease resistance protein [Populus trichocarpa] |
| 2 |
Hb_005595_020 |
0.2864141343 |
- |
- |
- |
| 3 |
Hb_072775_010 |
0.3046083404 |
- |
- |
60S ribosomal L37-3 -like protein [Gossypium arboreum] |
| 4 |
Hb_001916_080 |
0.3133991581 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 5 |
Hb_007484_040 |
0.3211288442 |
- |
- |
- |
| 6 |
Hb_016777_030 |
0.3331953929 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 7 |
Hb_157340_010 |
0.3413595314 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 8 |
Hb_000025_130 |
0.3506749448 |
- |
- |
- |
| 9 |
Hb_175069_010 |
0.354075822 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |
| 10 |
Hb_171529_010 |
0.3599820639 |
- |
- |
- |
| 11 |
Hb_015609_020 |
0.3683589492 |
- |
- |
PREDICTED: uncharacterized protein LOC105642775 [Jatropha curcas] |
| 12 |
Hb_000803_130 |
0.3686767385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000705_330 |
0.3730014885 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 14 |
Hb_003695_010 |
0.3732682215 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 15 |
Hb_003964_090 |
0.3765258582 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 16 |
Hb_002046_030 |
0.3768656586 |
- |
- |
PREDICTED: uncharacterized protein LOC104788746 [Camelina sativa] |
| 17 |
Hb_005839_050 |
0.3790293458 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 18 |
Hb_000347_510 |
0.3839344941 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 19 |
Hb_013818_050 |
0.389162958 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 20 |
Hb_013691_030 |
0.394658802 |
- |
- |
hypothetical protein CISIN_1g020131mg [Citrus sinensis] |