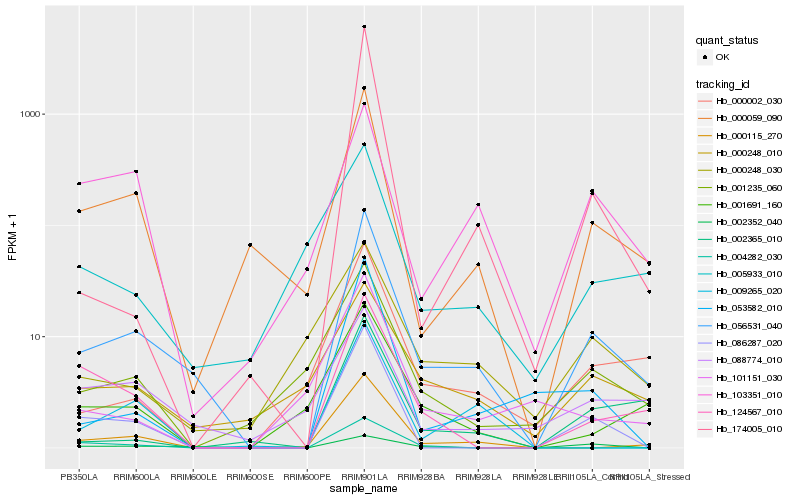
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056531_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103494239 [Cucumis melo] |
| 2 |
Hb_000059_090 |
0.1938239032 |
- |
- |
- |
| 3 |
Hb_000115_270 |
0.2063012036 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s13290g [Populus trichocarpa] |
| 4 |
Hb_086287_020 |
0.2143396233 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000002_030 |
0.2260774407 |
- |
- |
- |
| 6 |
Hb_124567_010 |
0.23538769 |
- |
- |
- |
| 7 |
Hb_053582_010 |
0.236165056 |
- |
- |
- |
| 8 |
Hb_002365_010 |
0.239752229 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 9 |
Hb_001235_060 |
0.2402192645 |
- |
- |
- |
| 10 |
Hb_000248_010 |
0.2402830002 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 11 |
Hb_088774_010 |
0.2454843581 |
- |
- |
- |
| 12 |
Hb_001691_160 |
0.2475491105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002352_040 |
0.2557452293 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 14 |
Hb_174005_010 |
0.2557912851 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 15 |
Hb_005933_010 |
0.2563647667 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 16 |
Hb_009265_020 |
0.2602918525 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000248_030 |
0.2605641389 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 18 |
Hb_101151_030 |
0.2642887991 |
- |
- |
- |
| 19 |
Hb_103351_010 |
0.2677898832 |
- |
- |
hypothetical protein POPTR_0002s01570g [Populus trichocarpa] |
| 20 |
Hb_004282_030 |
0.2716398205 |
- |
- |
hypothetical protein EUGRSUZ_B01857 [Eucalyptus grandis] |