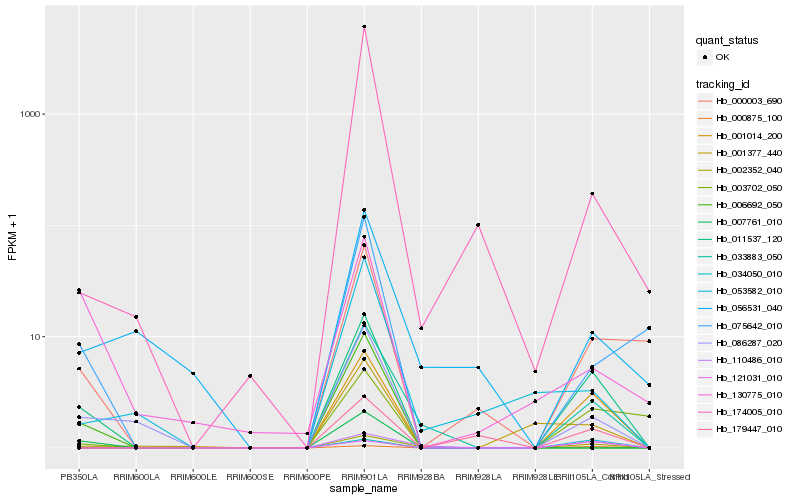
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_001014_200 |
0.1885421261 |
- |
- |
- |
| 3 |
Hb_086287_020 |
0.2265761237 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000875_100 |
0.2525967471 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 5 |
Hb_033883_050 |
0.2526121362 |
- |
- |
- |
| 6 |
Hb_000003_690 |
0.2561734798 |
- |
- |
- |
| 7 |
Hb_110486_010 |
0.2854893836 |
- |
- |
- |
| 8 |
Hb_121031_010 |
0.296123386 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 9 |
Hb_075642_010 |
0.2964391152 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 10 |
Hb_179447_010 |
0.297082002 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 11 |
Hb_003702_050 |
0.2974315572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 12 |
Hb_034050_010 |
0.3041519564 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 13 |
Hb_174005_010 |
0.304869709 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 14 |
Hb_001377_440 |
0.3115747728 |
- |
- |
- |
| 15 |
Hb_053582_010 |
0.3150276341 |
- |
- |
- |
| 16 |
Hb_002352_040 |
0.3156641918 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 17 |
Hb_130775_010 |
0.316250561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_056531_040 |
0.3207831483 |
- |
- |
PREDICTED: uncharacterized protein LOC103494239 [Cucumis melo] |
| 19 |
Hb_006692_050 |
0.321975841 |
- |
- |
Chloroplastic lipocalin [Theobroma cacao] |
| 20 |
Hb_007761_010 |
0.3254123267 |
- |
- |
- |