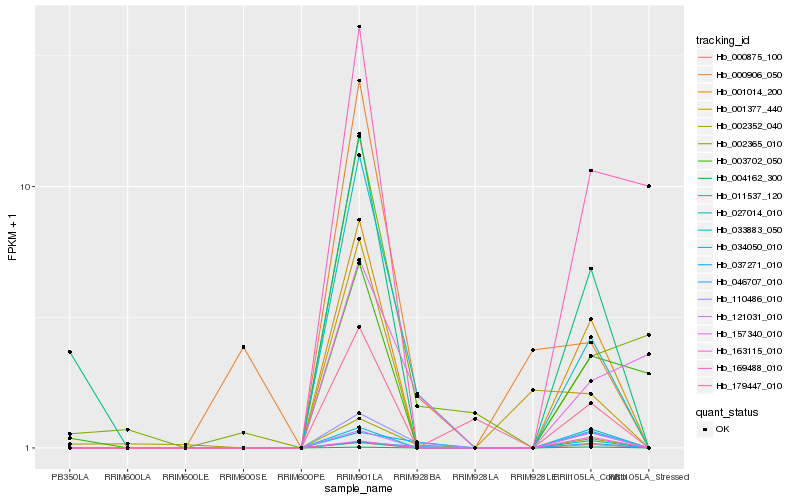
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110486_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_046707_010 |
0.1342048109 |
- |
- |
PREDICTED: uncharacterized protein LOC104887079 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_033883_050 |
0.1870472191 |
- |
- |
- |
| 4 |
Hb_037271_010 |
0.2062961214 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 5 |
Hb_001014_200 |
0.2129364335 |
- |
- |
- |
| 6 |
Hb_000875_100 |
0.2306624743 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 7 |
Hb_121031_010 |
0.2610066961 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 8 |
Hb_034050_010 |
0.2672326344 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 9 |
Hb_011537_120 |
0.2854893836 |
- |
- |
- |
| 10 |
Hb_004162_300 |
0.3112949218 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 11 |
Hb_027014_010 |
0.3169173059 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_179447_010 |
0.321740414 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 13 |
Hb_002352_040 |
0.3327041899 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 14 |
Hb_157340_010 |
0.3367824324 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 15 |
Hb_163115_010 |
0.3462404797 |
- |
- |
- |
| 16 |
Hb_169488_010 |
0.3526861547 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_003702_050 |
0.3544557097 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 18 |
Hb_000906_050 |
0.3549014147 |
- |
- |
- |
| 19 |
Hb_001377_440 |
0.358599109 |
- |
- |
- |
| 20 |
Hb_002365_010 |
0.3587765152 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |