Hb_001377_440
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig1377: 403543-403894 |
| Sequence |   |
Annotation
kegg
| ID | - |
|---|---|
| description | - |
nr
| ID | - |
|---|---|
| description | - |
swissprot
| ID | - |
|---|---|
| description | - |
trembl
| ID | - |
|---|---|
| description | - |
Gene Ontology
| ID | - |
|---|---|
| description | - |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_10445: 404394-407490 |
|---|---|
| cDNA (Sanger) (ID:Location) |
- |
Similar expressed genes (Top20)
Gene co-expression network
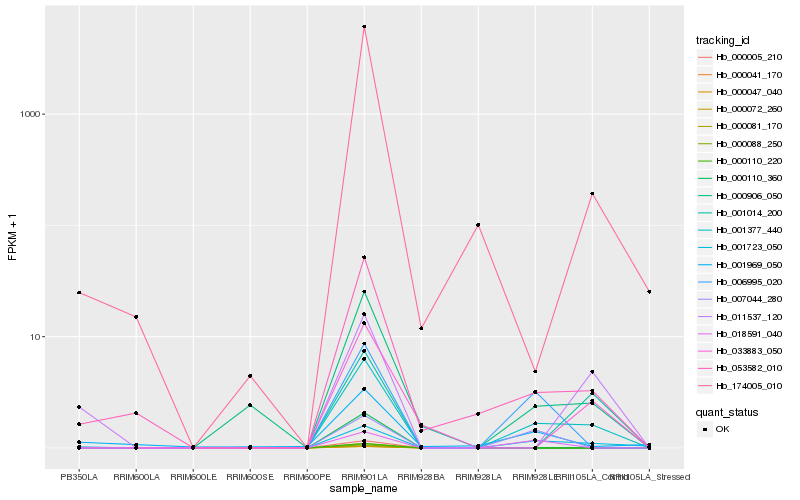
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 5.31332 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 0.610639 | 0 | 0 | 0 | 0.66384 |

